An automated microfluidic platform for investigating mutation accumulation
Imagine a tiny but powerful system that accelerates mutation accumulation experiments. This paper highlight presents a platform that allows researchers to simultaneously analyze multiple yeast strains. It's not just about speed, it creates possibilities for identifying mutagenic compounds and modeling diseases.
A Université Paris-Saclay, CEA, CNRS Paper
Paper: Sipos, E. H.; Léty-Stefanska, A.; Denby Wilkes, C.; Soutourina, J.; Malloggi, F. Microfluidic Platform for Monitoring Saccharomyces Cerevisiae Mutation Accumulation. Lab Chip 2021, 21 (12), 2407–2416. https://doi.org/10.1039/D1LC00086A.
This paper, published in Lab-on-Chip (2021), is a joint research work between the Interdisciplinary Laboratory on Nanoscale and Supramolecular Organization (LIONS) and the Institute for Integrative Biology of the Cell (I2BC) from the CEA/CNRS/Université Paris-Saclay.

The LIONS investigates nanostructured materials for practical applications in energy, environment, and health. They focus on safe and eco-friendly synthesis methods using statistical physics and advanced instrumentation, generating new knowledge and fostering commercialization through patents, licenses, and start-ups. Through a microfluidic team led by Dr. Florent Malloggi, their research work in microfluidics addresses a variety of challenges in fundamental chemistry, interface physics, and designing microfluidic chips for medical diagnostics and biotechnology.
The I2BC carries out research in the integrative biology of the cell around four thematic axes (expression and evolution of genomes, cell compartment and transport, stress and adaptation, and metabolism and bioenergy). Led by Dr. Julie Soutourina, the Genome Transcriptional Regulation (GTR) team from I2BC studies how genes are regulated at a genomic level, with a focus on the Mediator complex’s role in RNA polymerase II regulation. They use yeast and human cells to understand how transcription and other nuclear processes like DNA repair are coordinated.
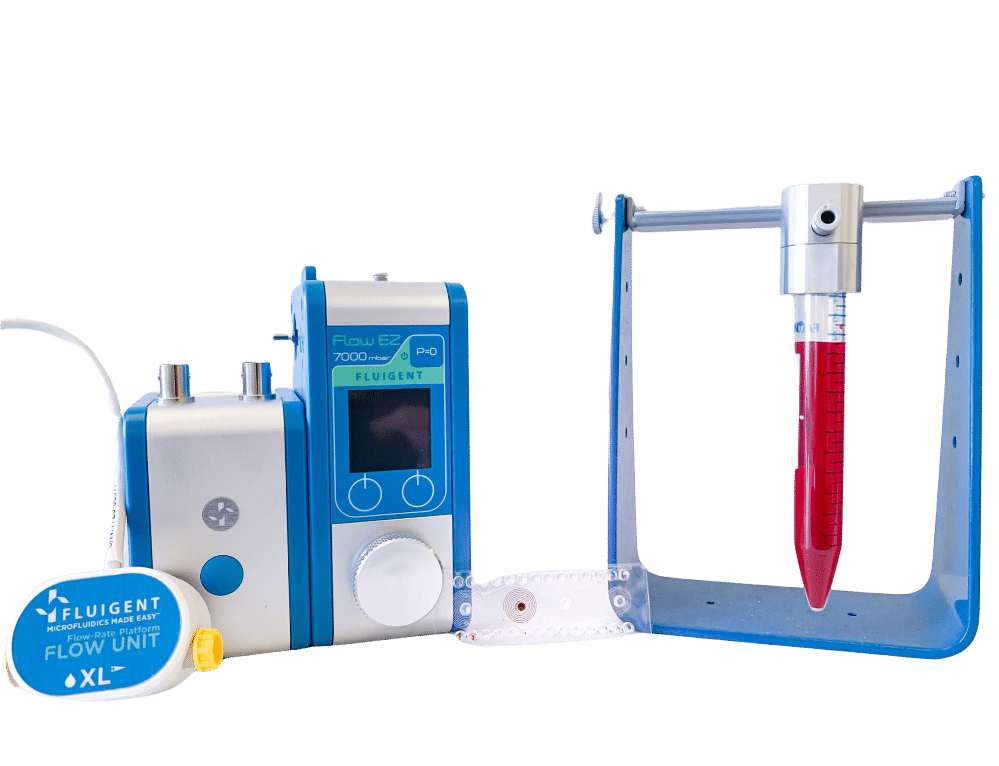
The LIONS and I2BC teams combined their expertise in microfluidics and genome understanding to develop an automated microfluidic platform, including Fluigent’s Flow EZ, for investigating mutation accumulation (MA). This approach highly reduced the experiment time and human intervention compared to traditional techniques.
Why is studying DNA mutations fundamental for advancing our understanding of genetics and human health?
Mutations serve as the driving force behind genetic diversity. They are necessary for evolutionary processes and also linked to diseases, notably cancer. In fact, mutations can arise naturally as cells divide throughout a person’s life. It is believed that a few dozen mutations occur during each cell division when DNA replicates.
On the other hand, DNA can undergo changes due to environmental factors, such as exposure to UV radiation, smoking, asbestos, or specific viruses. Therefore, it is vital to understand how mutations occur on a genomic scale.1,2 Nonetheless, studying concealed mutation events and their outcomes presents a challenge, as natural selection often obscures them from a biologists’ view.
How can we study hidden mutations and their effects without the impact of natural selection?
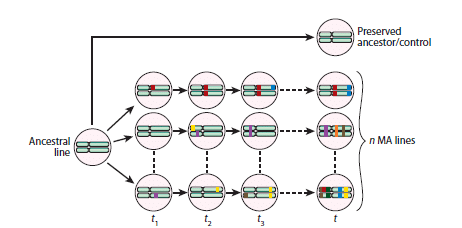
One of the main experiments used to study these mutation rates is mutation accumulation. It involves the expansion of a cell population from a single common ancestor under controlled conditions (such as in the presence of a molecule or when the ancestor’s genome contains a mutation of interest).
After several generations, a single daughter cell is unbiasedly isolated (creating a bottleneck population, thus minimizing natural selection), which then becomes the exclusive ancestor of a cell population that multiplies over numerous generations, continuing the cycle.
When complemented with high-throughput genome sequencing, this approach enables the precise mapping of spontaneous mutations, and the quantification of their occurrence rate, while also characterizing their effects on the phenotype.3,4
How are mutation accumulation experiments traditionally done?
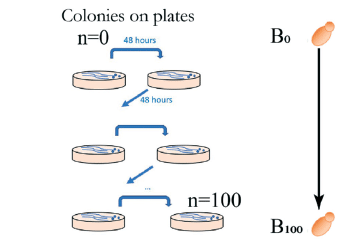
As the mutation rates are typically very low, classical mutation accumulation experiments are labor-intensive and time-consuming. They involve growing cells through a significant number of repeated plating on agar plates and introducing bottlenecks by randomly selecting a single colony at each step, a process that takes, for instance, two days for budding yeast at 30 °C.
This means that more than six months (and 800 Petri dishes) is required to achieve 100 single-cell bottlenecks in yeast mutation accumulation.
Additionally, the human intervention every 48 hours (the time it takes for individual yeast colonies to form) introduces bias, as the choice of a colony may not be entirely random and could be a non-random choice of mutants with higher fitness.
Also, human intervention can alter the cell growth environment by introducing bacterial or fungal contamination.
How can microfluidics automate mutation accumulation experiments?
Since its emergence in the early 1990s, microfluidics has automated laboratory experiments, offering advantages like high throughput, minimal material consumption, reduced cross-contamination, and small sample requirements.6
With this perspective, the researchers from LIONS and I2BC developed an automated microfluidic-based platform for mutation accumulation experiments based on budding yeast over many generations inside a specific microfluidic device.
Coupled with high-throughput sequencing, this approach simplifies and accelerates the process of measuring mutational profiles at the genome level with minimal human intervention.
How to set up a microfluidic platform for mutation accumulation
First, the cells chosen for the study were yeast Saccharomyces cerevisiae as they are an excellent unicellular eukaryotic model. In addition, they possess a compact genome and a short generation time compared to human cells.
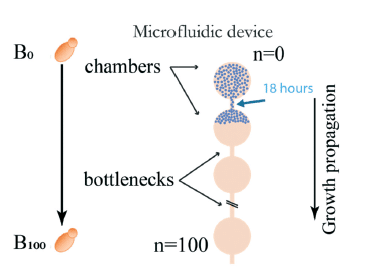
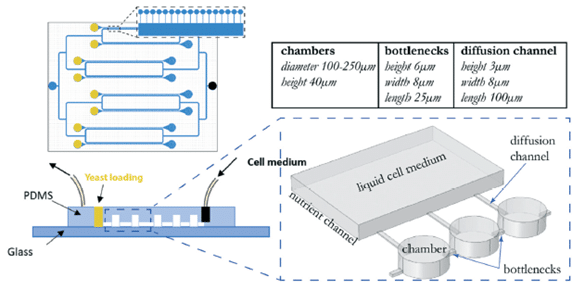
The strategy behind the microfluidic device is based on linear arrays of culture chambers where yeast populations grow for many generations. A narrow channel acts as a bottleneck, allowing only one cell to pass to a neighboring chamber. This single daughter cell then replicates until it occupies the entirety of the second chamber.
Afterward, another single daughter cell proceeds to the following chamber, and this cycle continues. This setup mimics the single colony-forming units in manual experiments. In one microfluidic device, 8 parallel channels enable 8 MA lines simultaneously.
To maintain growth for extended periods, nutrient channels connected by small diffusion channels are incorporated into the design, with Fluigent’s pressure-driven controller FlowEZ and FlowUnit regulating nutrient flow (8 µL/min) into the device.
Yeast growth was monitored using light microscopy, and after growth, yeast cells were collected for genome sequencing. To validate this microfluidic approach, the accumulation of mutations was done on two different genetic backgrounds, a wild-type strain and a base-excision DNA repair mutant characterized by a well-defined mutational profile.
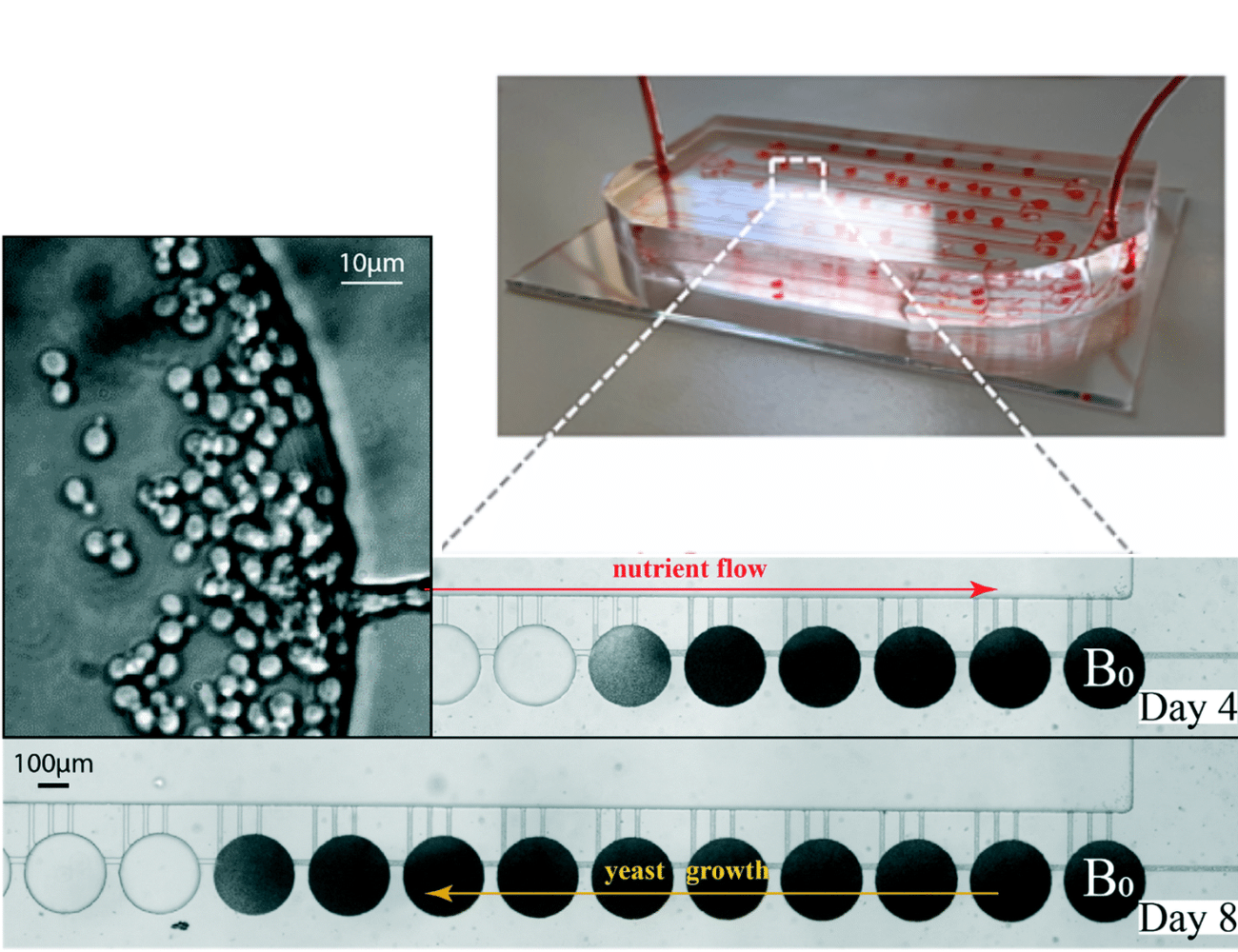
Proof of concept: Enhancing mutation accumulation experiments with minimal human intervention and time.
Through time-lapse imaging, it is possible to precisly follow the cell divisions and growth of yeast within the microfluidic device and their random movement into the following chambers. When a yeast cell reached a new chamber, it endured division and took over the chamber as demonstrated in the video, thus confirming the efficiency of the chip design.
The stable and precise nutrient flow, set up through the Flow EZ and Flow Unit, ensured long-term cell culturing.
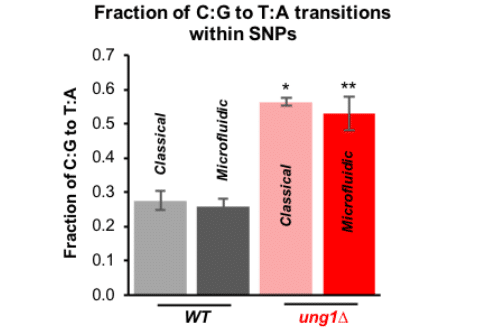
The high-throughput genome sequencing results also confirmed that the microfluidic device successfully detected the specific mutational signature and enabled accumulation of mutation in a way comparable to the classical method (figure 6).
This approach presents several advantages compared to the classical mutation accumulation method:
- Reducing time and number of steps, which traditionnaly involves 800 Petri dishes and frequent human intervention over a period of more than 6 months. A single microfluidic chip with up to 8 mutation accumulation lines running in parallel required 3 months and minimal intervention, typically once every two weeks, to ensure the device’s proper functioning.
- Automating the hand-picking step of a single colony, thus eliminating the bias associated with human intervention and consequent contaminations.
- Providing a controlled growth environment in terms of temperature, and flow of nutrients.
Conclusion
In this paper highlight, E.Sipos et al., from the LIONS and I2BC research groups (CEA-CNRS-Université Paris-Saclay), developed a microfluidic device that streamlined mutation accumulation experiments, reducing the time required from over 6 months to 1-4.5 months. It allows parallel analysis of multiple yeast strains, facilitating unbiased genome-wide mutational process comparisons.
The device holds promise for identifying mutagenic compounds and discovering new therapeutic targets. It may also help in developing human cell culture systems to replicate mutation accumulation in diseases like cancer.
Related products
Related Resources
- Expert Reviews: Basics of Microfluidics
Flow control for droplet generation using syringe pumps and pressure-based flow controllers
Read more - Expert Reviews: Basics of Microfluidics
Micropipette aspiration of cells and tissues
Read more - Expert Reviews: Basics of Microfluidics
Prostate Organoid Culture in Microbeads
Read more
References
- (1) P. L. Foster, in Methods in Enzymology: DNA Repair, Part B, 2006, vol. 409, pp. 195–213.
- (2) A. Frenoy and S. Bonhoeffer, PLoS Biol., 2018, 16, e2005056.
- (3) J. E. Barrick and R. E. Lenski, Nat. Rev. Genet., 2013, 14,827–839.
- (4) V. Katju and U. Bergthorsson, Genome Biol. Evol., 2019, 11,136–165
- (5) D.L. Halligan andP.D. Keightley, Annu. Rev. Ecol. Evol. Syst. 2009, 40 (1), 151–172.
- (6) L. Y. Yeo, H. C. Chang, P. P. Y. Chan and J. R. Friend, Small,2011, 7, 12–48.