High Throughput Single Cell Analysis
Unlocking the Hidden World of Individual Cells: Single-Cell Analysis
Individual cell heterogeneity within a population has invalidated historic classification methods based on macroscopic considerations and given rise to new evaluation techniques based on single cell transcriptional signatures. In this context, droplet microfluidics has emerged as a powerful tool for single cell analysis and manipulation thanks to its high throughput screening capacities, easy fluid handling, and reduced costs related to device miniaturization.
What is Single Cell Analysis?
Most cellular research from fundamental cell biology and microbiology to applications in biotechnology is performed using cell populations with high cell numbers. This is convenient, as a huge variety of experimental techniques exist to analyze cell behavior. However, cell population data only covers information of an imaginary average cell (i.e. the average of the microbial culture), and not the mechanistic information of one cell. (1)
The Significance of Single-Cell Analysis:
Recent research provides substantial evidence that the heterogeneity of individual cells within genetically identical populations can have a profound impact on their survival.
Methods that use average responses from a population often mask the difference from individual cells. To fully understand cell-to-cell variability, a complete analysis of an individual cell, from its live state to cell lysates, is essential (1,2).
Distinguishing Bulk vs. Single-Cell Approaches:
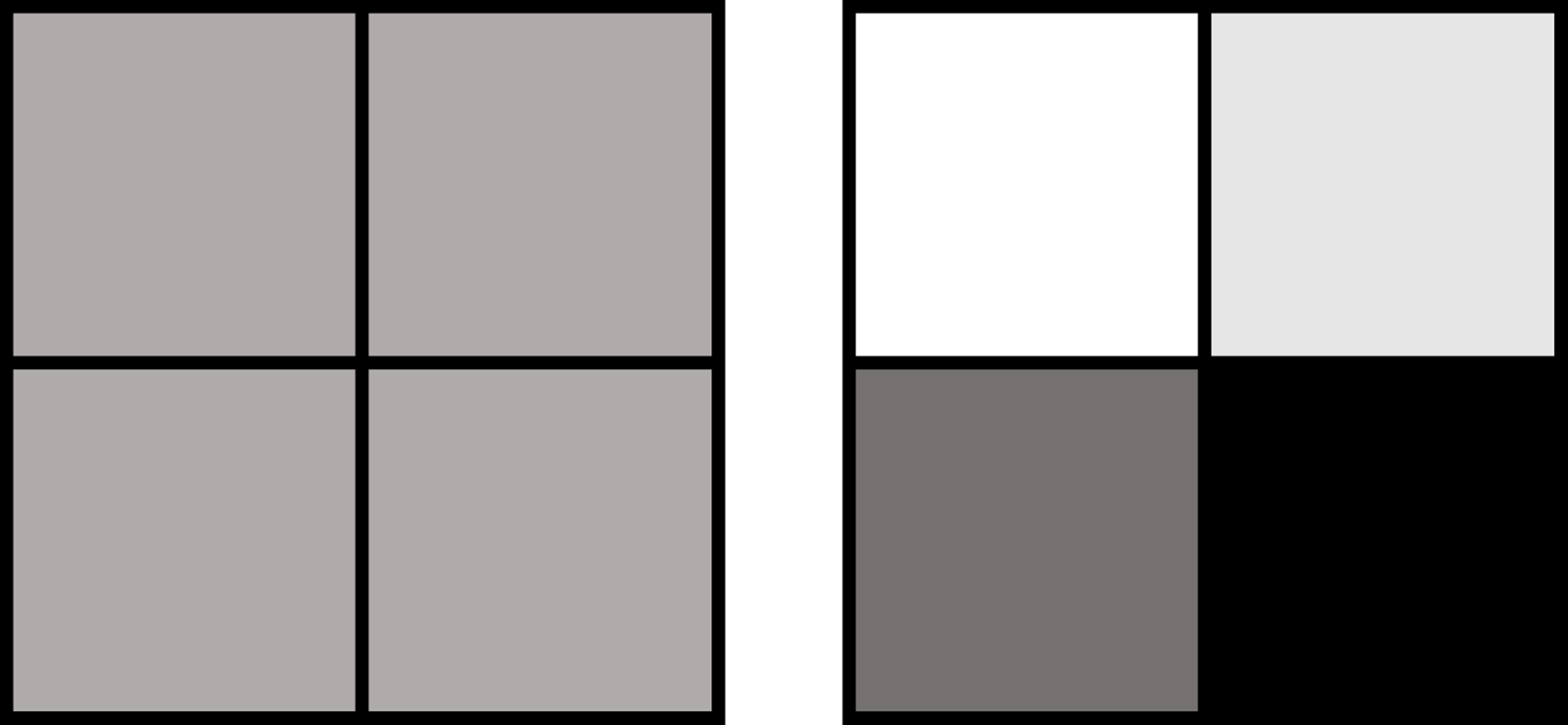
To understand the distinction between the bulk and the single-cell approaches, imagine you have four squares: each of them is colored with a different shade of grey (figure 1). With the single-cell approach, you will see the squares as they are, with their actual colors, whereas with the more classic approach you will see four identical grey squares, hiding the color differences between each square.
Unveiling the Secrets of Single Cell Properties and Analysis Technologies
Within a population, the unique properties of single cells can be quantified using various cutting-edge technologies like flow cytometry and Elispot. These techniques enable the exploration of cell-to-cell variations, unveiling the heterogeneity within the population.
However, it’s essential to recognize that this analysis captures only a snapshot, offering insights into a cell’s state at a specific moment. It doesn’t provide a historical perspective or track a cell’s temporal development.
Moreover, in cases where the analysis is single-cell, the bioassay itself is bulk, with cells interacting with one another, potentially leading to changes in individual responses to stimuli. A cell can initiate its response upon detecting an activation signal from another cell.
Advancements and the Pursuit of Spatiotemporal High-Throughput Analysis:
Beyond these conventional endpoint technologies, the field is experiencing rapid developments with the ultimate goal of achieving spatiotemporal high-throughput single cell analysis, enabling a mechanistic understanding of cellular functions. The enthusiasm in this field is evident in the exponential growth of single cell studies and conferences in recent years.
This progress is driven by technical advancements in microfluidic chip manufacturing and analytical methods, as well as the increasing demand from biologists (3).
Key Challenges and the Role of Microfluidics and Lab-on-a-Chip Technology:
The sensitive detection of multiple cellular components and high-throughput analysis of individual cells pose significant challenges in achieving this goal. In this context, microfluidics and lab-on-a-chip technology have emerged as the most promising approaches to address these challenges. These innovative technologies hold the potential to unlock new insights into single cell properties and their roles within populations.
Single‐Cell Analysis Using Droplet Microfluidics
Recent analysis of healthy and diseased tissue homogeneous at the macroscopic scale revealed striking heterogeneities at cellular levels. This variability is particularly well illustrated in polyclonal tumors which constantly undergo mutations.
In this respect, single cell analysis is necessary to fully capture the complexity of such tissue. However, working at cellular scale equally exposes many variations in gene expression: from specific biomarkers to insignificant delays in gene expression. High throughput analysis is then needed to multiply the number of profiled cells and discriminate relevant biomarkers from intrinsic population noise.
Droplet microfluidics is particularly well suited and extensively used for high throughput single cell analysis: individual cells are isolated and confined at high speed in pico-volumes to analyze biological processes at the cellular level, streamlining multiple procedures on a single chip, with scope for parallelization.
Recently developed droplet microfluidics has also emerged as a new forerunner for single-cell encapsulation and analysis with massive parallelization. High throughput screening of rare cells to a drug library has been achieved, providing additional information on cell heterogeneity response. The use of microdroplet confinement has enabled new insights into the nature of quorum sensing, suggesting it is a “cell-autonomous mechanism for diffusion or efficiency sensing”.
Why is droplet microfluidics well-suited for single-cell analysis ?
High monodispersity: unique liquid-handling capabilities of microfluidic systems
Reduced costs: volume down scaling from µL to pL compared to pipetting robots
Time saving: molecular diffusion length reduced in small volumes
Higher sensitivity: smaller molecular quantity (1.106-fold less) required to reach minimum detectable concentration
High throughput: up to 1.106 cells compartmentalized per second.
Applications
Drop-Seq:
Single-cell genome or transcriptomic sequencing aims to increase our understanding of complex microbial ecosystems and disease in multicellular organisms by isolating the contributions of distinct cellular populations.
Unlocking the Potential of Flow Cytometry:
Droplet microfluidics technology has opened the door to groundbreaking capabilities in flow cytometry and (FACS). It allows for the encapsulation of single cells within double emulsions, transforming them into miniature microbioreactors. Within these microreactors, the fluorescence signals produced by specific molecules are captured and analyzed, enabling precise assessment of protein and metabolite secretion.
Through this innovative approach, flow cytometry gains the ability to delve into the intricate world of protein and metabolite secretion. The double emulsions serve as microscale laboratories, where the fluorescence signals arising from specific molecule production can be effectively trapped and scrutinized.
Sorting with FACS for Specific Cell Types:
Beyond flow cytometry, the integration of fluorescence-activated cell sorting (FACS) technology empowers researchers to selectively collect specific cell types from complex populations. This technology facilitates the precise separation and retrieval of cells of interest, providing invaluable insights for a wide range of applications.
Microbiome culture:
Compartmentalization of single bacteria inside droplets makes it possible to start new cultures inside each droplet, preventing growth competition between the different strains and allowing even rare bacteria to grow and form their own culture
Cell sorting with microfluidics:
Single cell sorter microfluidic platforms provide numerous advantages over conventional methods by reducing the size of necessary equipment, eliminating potentially biohazardous aerosols, and simplifying the complex protocols commonly associated with cell sorting.
Personalized medicine:
Single cell analysis of a tumor’s heterogeneity associated with selection and amplification of specific corresponding T cells for personalized cancer immunotherapy.
Selected publications from our customers:
Yin, H. and Marshall, D. (2012) “Microfluidics for single cell analysis,” Current Opinion in Biotechnology, 23(1), pp. 110–119. Available at: https://doi.org/10.1016/j.copbio.2011.11.002.
Andersson, H. and van den Berg, A. (2004) “Microtechnologies and nanotechnologies for single-cell analysis,” Current Opinion in Biotechnology, 15(1), pp. 44–49. Available at: https://doi.org/10.1016/j.copbio.2004.01.004.
Yin, H. and Marshall, D. (2012) “Microfluidics for single cell analysis,” Current Opinion in Biotechnology, 23(1), pp. 110–119. Available at: https://doi.org/10.1016/j.copbio.2011.11.002.
Related Products
Related resources
WEBINAR: Single cell encapsulations compatible with FACS sorting, API encapsulations in biocompatible polymers, and more
Discover- Microfluidic Application Notes
A quick and efficient double encapsulation method for FACS-based droplet sorting
Discover - Microfluidic Application Notes
What is the best method for Microencapsulation of Bacteria and Yeast in Small Double Emulsions?
Discover Flow Expertise for Cell Encapsulation and Single-Cell Analysis
DiscoverWEBINAR – Master the production of Double Emulsions
Discover- Microfluidic Application Notes
Encapsulation of Cells In Small Double Emulsions
Discover - Microfluidics White Papers
Double emulsion for the generation of microcapsules – a Review
Discover - Microfluidic Application Notes
Double Emulsion Generation
Discover A complete cell sorting pack for starting size sorting experiments
Discover