The Hebrew University: Encapsulation and culture in 3D hydrogels for Single cell sequencing
The Epigenomics Ram Laboratory, part of the Department of Biological Chemistry of Alexander Siberman Institute of Life Sciences at The Hebrew University in Jerusalem, Israel. The Ram Laboratory’s mission is to characterize chromatin mechanisms and transcriptional output at the single-cell level to better understand cellular processes in health and disease. To do so, they develop and utilize cutting-edge technology including microfluidics, molecular and cellular biology, and computational biology.
More specifically, they adapt RNA-seq and ChIP-seq methods at the single-cell level using droplet-based microfluidic devices, such as Fluigent’s pressure-driven flow controllers along with a dedicated software for microfluidic applications. CloneSeq combines clonal expansion inside three-dimensional (3D) hydrogel spheres and droplet-based single cell sequencing and allows for the identification of the presence of novel cancer-specific subpopulations.
“Microfluidics is a robust and cost-effective technology for single cell and single clone processing”
“Fluigent made microfluidics parallelization a feasible task”
CloneSeq: A highly sensitive single-cell analysis platform for the comprehensive characterization of cells from 3D culture
Introduction
Single-cell studies have revealed that there is considerable cell-to-cell variation within tumors of different cancer types and during embryonic stem cell (ESC) differentiation 1–4. However, in many cases, single cell experimental data is still difficult to interpret as a high degree of randomness persists.
To overcome this limitation, Bavli et al. developed a complementary single cell sequencing technology: CloneSeq. This method combines clonal expansion inside three-dimensional (3D) hydrogel spheres and droplet-based RNA sequencing (RNA-seq)5. The authors revealed the presence of novel cancer-specific subpopulations, including cancer stem-like cells, which are not identified using standard RNA sequencing assays.
Barcode formation, cell encapsulation in hydrogels, and single cell sequencing (including InDrops and Drop-seq) were all performed using Fluigent Flow-EZ pressure-based flow controllers, controlled with Fluigent OxyGEN software with pressures ranging from 69 mbar to 2 bar.

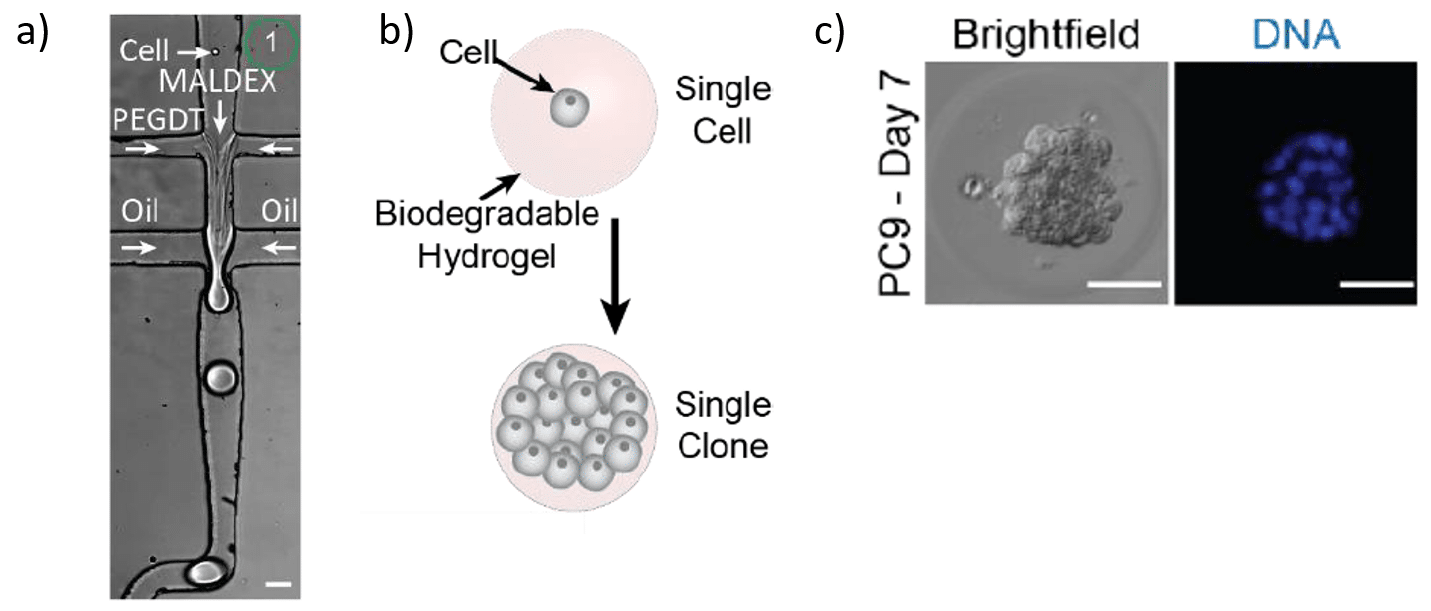
Step 1 – Single-cell encapsulation in hydrogels for 3D cell culture system
The first step of the CloneSeq method consists of capturing single cells inside a biodegradable hydrogel for subsequent clonal expansion. All flow rates are finely controlled using Fluigent Flow EZs combined with our Flow Units and OxyGEN software. Flow rates used range from 8 to 34 µL/min depending on the solution. About 700 hydrogel spheres per second are generated under these parameters5.
The cured gels are collected into a tube and immersed in culture medium, permitting the proliferation of the encapsulated cells and clone formation (figure 2).
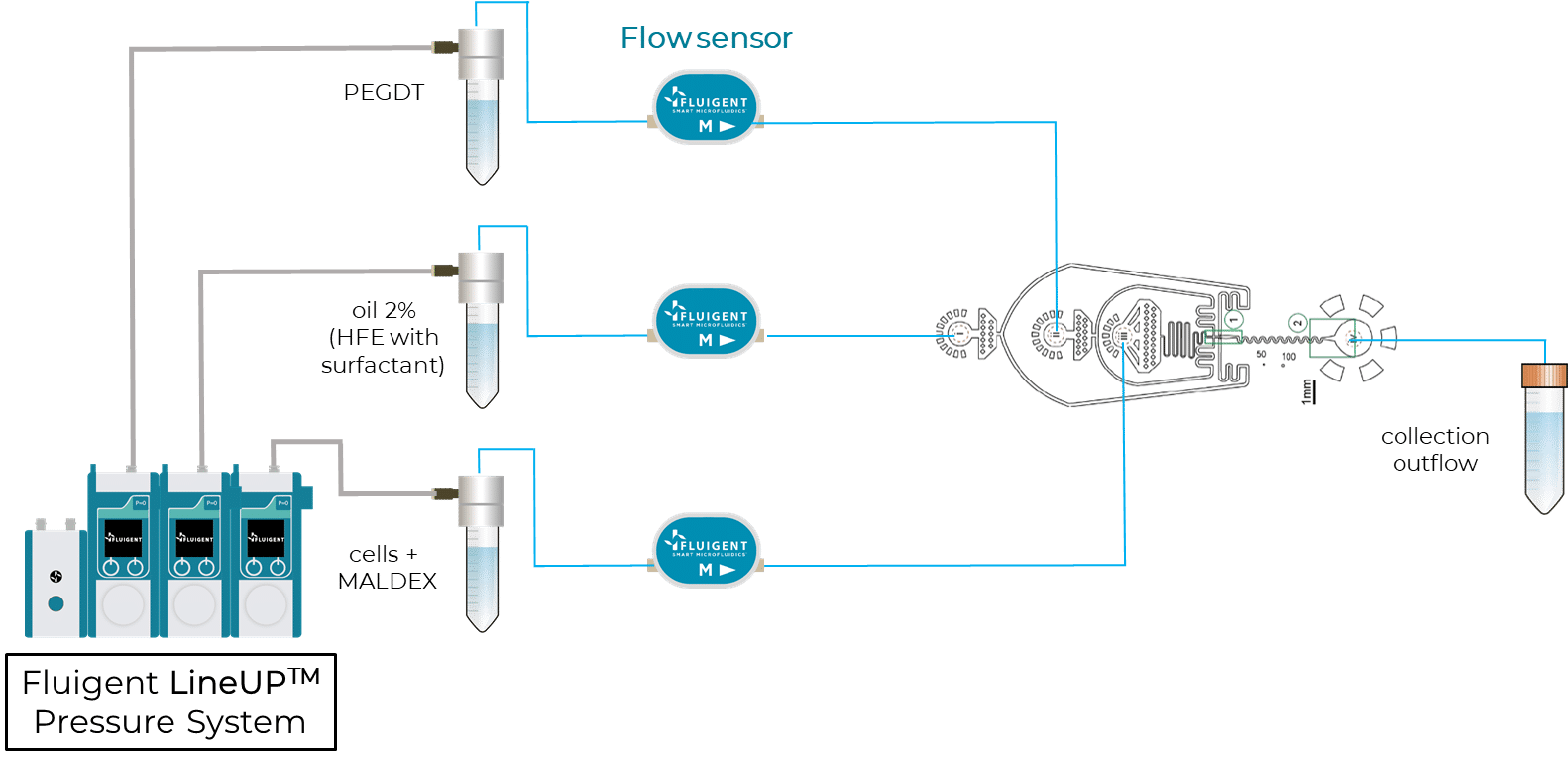
Step2 – CloneSeq: profiling of clones using modified InDrops and Drop-Seq protocols
For the RNA profiling of clones, the authors designed a microfluidic device to capture clones in droplets and barcode their mRNAs using a custom InDrops protocol5. Figure 3 shows a scheme of this InDrop-based microfluidic configuration. Once again, flow rates are finely controlled using Fluigent Flow EZs combined with our Flow Units and OxyGEN.
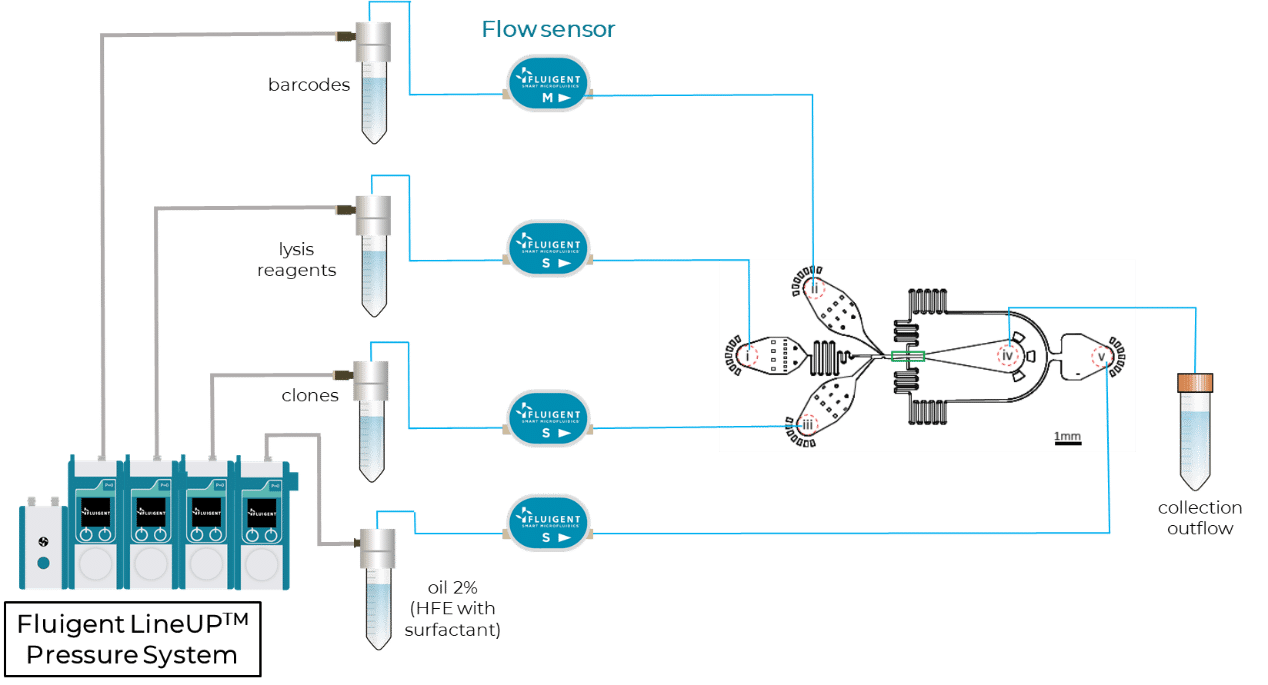
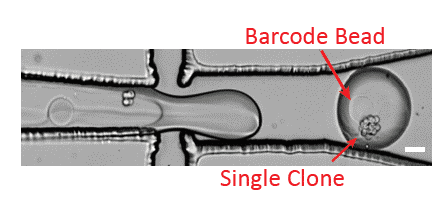
Partial results
To study the effect that clonal expansion within the hydrogel spheres has on the homogeneity of cell states within the clones with droplet-based single cell sequencing, the authors compared the inter-clone correlations of small (n<15 cells) and large (n≥15 cells) clones and of pseudo-clones. They showed for human PC9 cells that cells sharing a clonal origin are more similar to each other compared to random cells, suggesting that clones are homogeneous5.
Cleaning protocol
After the experiments, the authors perform an automated cleaning protocol using the Fluigent M-Switch, a microfluidic valve allowing for the injection of up to 10 fluids in a sequential manner.
Conclusion
With this case study, the authors demonstrated the use of Fluigent pressure-driven flow controllers along with a dedicated software to provide excellent flow control for cutting-edge microfluidic applications including single-cell encapsulation and culture within 3D hydrogels droplets, and for InDrops protocols followed by RNA sequencing.
Related Products
Related Resources
References
- Li, H. et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 49, 708–718 (2017).
- Dalerba, P. et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 29, 1120–1127 (2011).
- Kim, K. T. et al. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol. 16, 1–15 (2015).
- Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science (80-. ). 352, 189–196 (2016).
- Bavli, D. et al. CloneSeq: A highly sensitive analysis platform for the characterization of 3D-cultured single-cell-derived clones. Dev. Cell (2021).
Oren Lab website & other research articles
https://ramoren3.wixsite.com/oren-ram-lab
https://www.bio.huji.ac.il/en/departments_biological_chemistry_en
Esrrb is a cell cycle dependent priming factor balancing between pluripotency and differentiation