What is the best method for Microencapsulation of Bacteria and Yeast in Small Double Emulsions?
The precise manipulation, quick cultivation, and delicate detection of yeast and bacteria are crucial in microbiology and biomedicine for behavior monitoring, identification of phenotypes, physiological assessment, and molecular analysis (1, 2).
Microfluidic droplets have proven to be an effective method for encapsulating yeast and bacteria in highly monodisperse droplets for high-throughput screening and analysis of their phenotypes, subcellular structures, genes, and metabolites (3).This application note details the encapsulation of the yeast strain S. cerevisiae CEN.PK 113-7D and the bacterial strain L. cremorsi MG1363_GFP in double emulsions of 42 µm size using the Cell Encapsulation Platform, resulting in Microencapsulation of Bacteria and Yeast.
Advantages of bacteria and yeast microencapsulation method
In recent years, the microencapsulation of Bacteria and Yeast has expanded due to the advantages and novel information provided by these methods. In particular, droplet microfluidics has become a popular technique for bacterial and yeast encapsulation due to its performance, efficiency, and precision. This method has been used in various areas of microbiology, including pathogen detection and identification, antibiotic susceptibility testing, microbial physiology studies, and biotechnological applications (1).
Bacteria and Yeast encapsulation is a crucial technique in the investigation of microorganisms that play a vital role in the ecosystem and have a potent metabolic capacity for producing biopharmaceuticals and recombinant proteins (2,3).
Double WOW emulsion for cell encapsulation
To perform this encapsulation method, double water-in-oil-in-waterDouble Emulsion Generation (W/O/W) emulsions containing the microorganisms of interest are generated. Unlike single water-in-oil (W/O) emulsions, double emulsions provide an aqueous carrier fluid, making them compatible with most flow cytometry and cell sorting setups. They also protect sensitive biological structures from the sheath fluid, preserve the link between microorganisms and the substances they secrete, and allow for in vitro analysis of biomolecules (4).
Droplet-based microfluidic techniques, particularly for the microencapsulation of Bacteria and Yeast, offer superior control over droplet generation, producing highly stable and monodisperse double emulsions. To be compatible with cell sorting and analysis devices, the droplets must be small (< 60µm) in diameter and large enough to encapsulate the variants of interest (5).
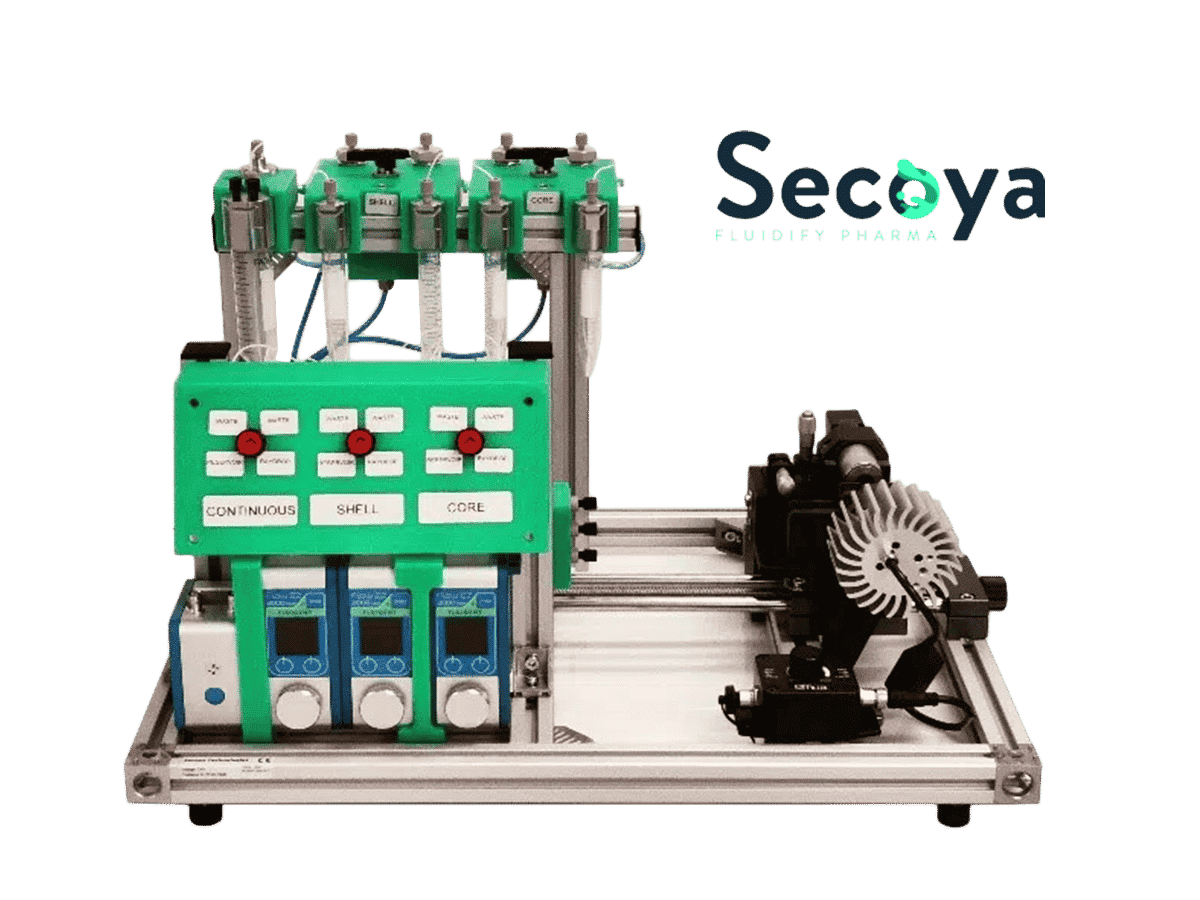
Therefore, in this application note, we demonstrate an easy-to-use and reliable workflow for encapsulating the yeast strain S. Cerevisiae CEN.PK 113-7D and the bacterial strain L. cremoris MG1363_GFP into highly monodisperse double emulsions with a diameter of 42 μm, suitable for high-throughput screening and sorting. The workflow uses the Cell Encapsulation Platform, consisting of Fluigent’s fluid handling system and Secoya’s emulsification technology, the RayDrop.
This application note was created in collaboration with Secoya and TU Delft. The images used are courtesy of TU Delft; Marijke Luttik, Sagarika B. Govindaraju and Rinke van Tatenhove-Pel.
How to encapsulate bacteria and yeast in small double emulsions
Materials
Yeast encapsulation
Reagents
Continuous phase:
• Water + 2% Tween20
Shell phase:
• dSurf (HFE7500 + 2% biocompatible surfactant)
Core phase:
• S. Cerevisiae CEN.PK 113-7D was grown on SM, a chemically defined medium for Yeast.
Bacterial encapsulation
Reagents
Continuous phase:
• Water + 2% Tween20
Shell phase:
• dSurf (HFE7500 + 2% biocompatible surfactant)
Core phase:
• Lactococcus lactis subsp. cremoris MG 1363 was grown on Chemically Defined Medium for prolonged cultivation (CDMPC).
Methods
- Prepare the shell and core solutions by filtering them using filters with pore sizes of 0.2 µm and 40 µm.
- Fill the appropriate reservoirs with the filtered shell and core solutions.
- Create a single emulsion of the shell solution by adjusting the flow rates of the continuous and shell phases.
- Initiate the encapsulation process by introducing the core solution into the single emulsion to form a double emulsion. This is done by adjusting the flow rate and directing the main phase valve in the reservoir towards the device used for emulsification (such as the RayDrop).
- After a few seconds, collect the encapsulated bacteria or yeast in Falcon tubes for further analysis. The double emulsions will have a protective shell surrounding the core solution, which contains the bacteria or yeast allowing for in vitro analysis.
Visit the Cell Encapsulation Platform datasheet and User Manual to learn more
Results
Once the Microencapsulation of Bacteria and Yeast method is complete, the generated double emulsions are visualized under a microscope to check their monodispersity and stability.
Yeast Encapsulation in Small Double Emulsions
Figure 1 shows a successful encapsulation of yeast (S. cerevisiae CEN.PK 113-7D) in DE droplets of 42 µm in size.
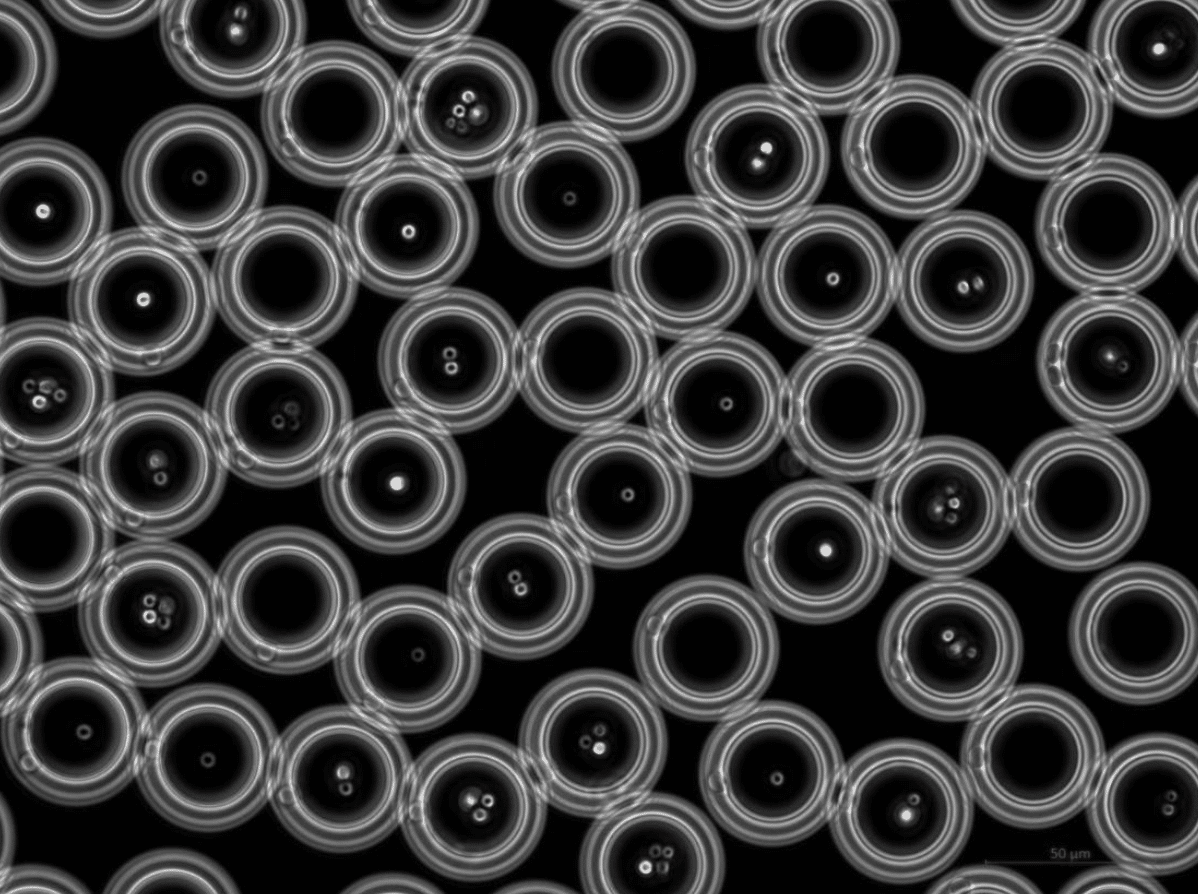
Figure 1. The yeast strain S. Cerevisiae CEN.PK 113-7D encapsulated within double emulsions of 42 µm.
Courtesy of TU Delft Marijke Luttik, Sagarika B. Govindaraju and Rinke van Tatenhove-Pel.
Bacterial Encapsulation in Small Double Emulsions
Figure 2 shows bacteria successfully encapsulated inside double emulsions with a diameter of 42µm. In both images, a high monodispersity of the droplets can be observed, as well as a homogeneous distribution of the biological material.
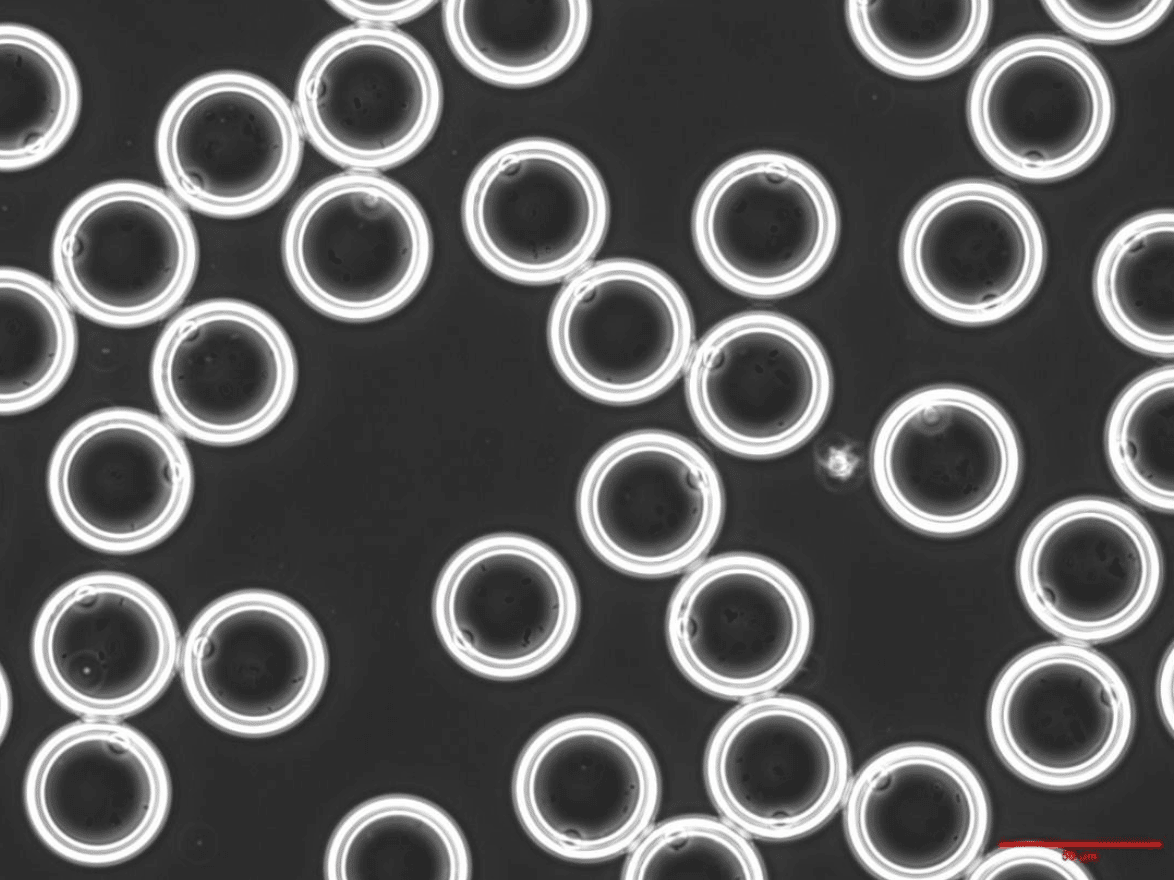
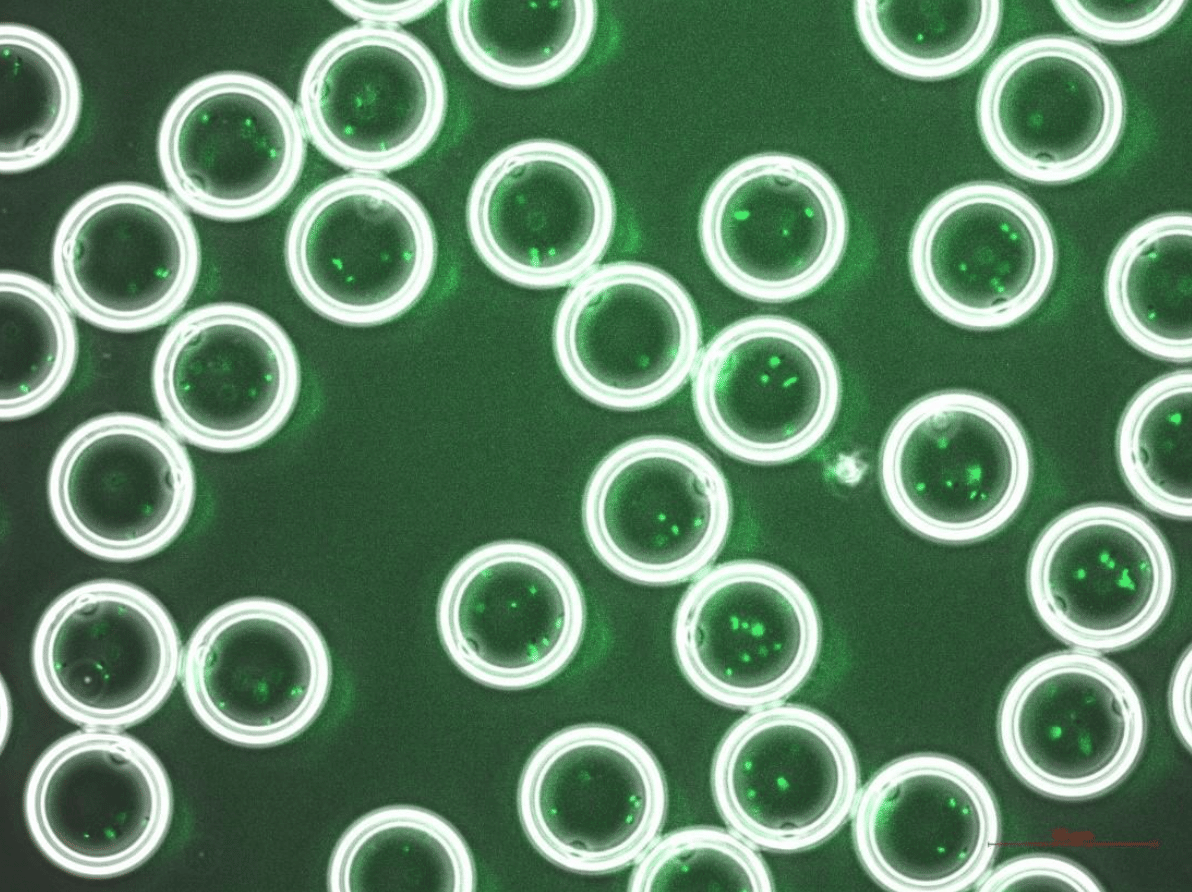
Figure 2. The bacterial strain L. cremoris MG1363_GFP, encapsulated within double emulsions of 42 µm in diameter. Bright field (up); Overlay of brightfield and fluorescence (down).
Courtesy of TU Delft Marijke Luttik, Sagarika B. Govindaraju and Rinke van Tatenhove-Pel.
Conclusion
In this application note, we demonstrate that the Cell Encapsulation Platform can achieve precise droplet size control in the Microencapsulation of bacteria and yeast in w/o/w double emulsions. We also show that the RayDrop is capable of creating monodisperse double emulsions with outer diameters of less than 60 µm, small enough for further analysis.
The yeast strain S.Cerevisiae CEN.PK 113-7D and the bacterial strain L.cremoris MG1363_GFP were successfully encapsulated in double emulsions with a diameter of 42 micrometers. The high monodispersity and stability are a result of Fluigent’s stable and pulse-free pumping technology and Secoya’s emulsification technology, the RayDrop. Other RayDrop configurations and nozzle dimensions are available to produce droplets of different sizes for various applications. This is due to the RayDrop’s versatility and flexibility, which allows users to easily adjust the configuration and droplet range produced.
WEBINAR: Encapsulation of fluorescent bacteria in double emulsions for FACS sorting
Optimize your FACS experiments by encapsulating single cells or bacteria inside 30µm double emulsions.
- Learn how to use our double emulsion platform to encapsulate bacteria in W/O/W DE
- Discover the DE sorting efficiency using FACS
- Have a live discussion with our experts concerning your specific needs
Related products
Expertises & Resources
-
Microfluidic Application Notes A quick and efficient double encapsulation method for FACS-based droplet sorting Read more
-
Microfluidics White Papers Double emulsion for the generation of microcapsules – a Review Read more
-
Microfluidic Application Notes Encapsulation of Cells In Small Double Emulsions Read more
-
Microfluidic Application Notes Double Emulsion Generation Read more
-
Microfluidic Application Notes Water in Oil Emulsions Read more
-
Expert Reviews: Basics of Microfluidics Microfluidic Droplet Production Method Read more
-
Expert Reviews: Basics of Microfluidics Flow control for droplet generation using syringe pumps and pressure-based flow controllers Read more