Analysis of a commercial surfactant for digital PCR assay
Digital PCR assay represents an example of the power of PCR and provides unprecedented opportunities for molecular genetic analysis in cancer. The technique is to amplify a single DNA template from minimally diluted samples, therefore generating amplicons that are exclusively derived from one template and can be detected with different fluorophores or sequencing to discriminate different allele. D
igital PCR study has been applied in quantification of mutant alleles and detection of allelic imbalance in clinical specimens, providing a promising molecular diagnostic tool for cancer detection. In this application note we are investigating the usability of the commercially available surfactant dSurf for an exemplary digital PCR-assay.
We kindly thank of Photonic Leibniz Institute Technology for this collaboration, and for sharing the results obtained with their system. For information about Photonic Leibniz Institute Technology : https://www.leibniz-ipht.de/

Introduction to digital PCR
What is digital PCR?
Digital droplet-based assays offer promising opportunities for the absolute quantitation of low concentration analytic species. During the last decade digital-PCR assay (dPCR) became one of the most prominent assays for this class of analytical methods.
For performing the assay, the sample volume is split into multiple droplets in such a way that each droplet contains either one or none of the target DNA molecules.
Due to the small droplet volume, the PCR reaction runs very efficiently even from a single molecule.
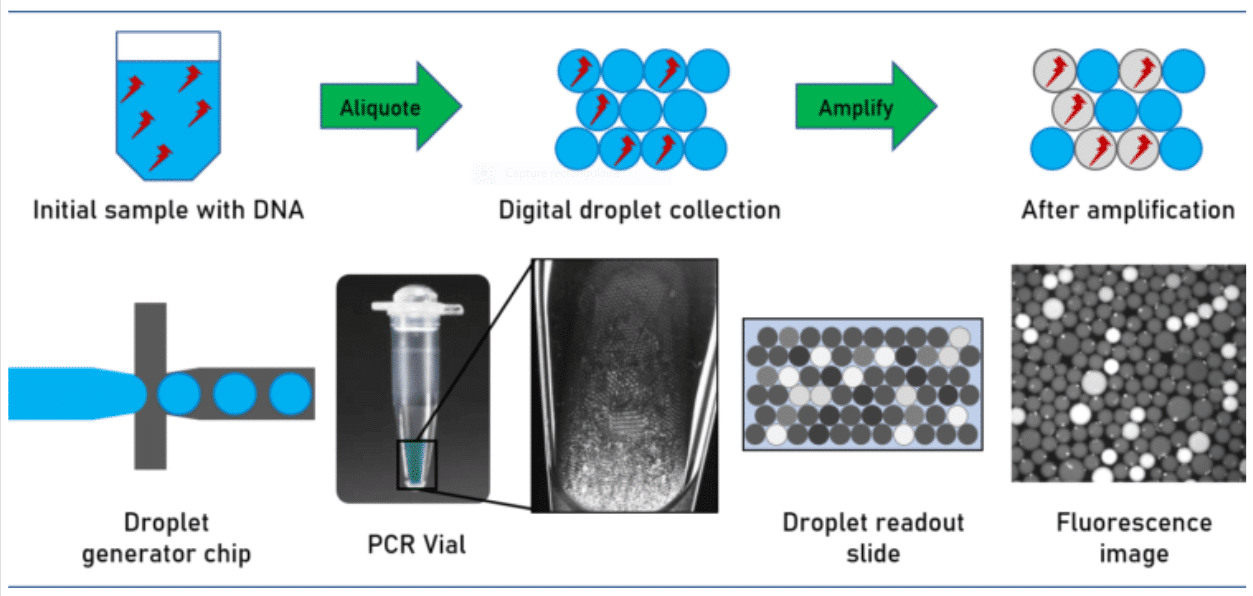
How does digital PCR work?
During amplification, a fluorescent dye is formed or activated. The positive droplets become fluorescent. Absolute quantitation of the number of target molecules is simplified to the count of fluorescence active droplets in the generated droplet collection. Not regarding the simplicity of the approach, its technical implementation is challenged by stabilizing the droplets collected over the complete assay avoiding unwanted droplet coalescence or crosstalk between the droplet ingredients. This has been solved by utilizing perfluorinated mineral oils as the carrier oil in combination with advanced perfluorinated surfactants, which stabilize the emulsion and avoid crosstalk and DNA exchange between the individual droplets.
In this application note we are investigating the usability of the commercially available surfactant dSurf for an exemplary digital PCR assay.
What are the materials and equipment for digital PCR experimentation?
The Fluigent droplet kit was employed for the experiments, utilizing the Fluigent EZ Drop chip with three microfluidic droplet generators per chip. Interconnection between the chip and fluid reservoirs was achieved using 2m PEEK 1/32″ tubing with an outer diameter of .010″ and two sleeves with 1/16″ outer diameter, .033″ inner diameter, and 1.6″ length. More details and dimensions of the droplet generation chip can be found on the Fluigent website.
Pressure-driven flow control was managed through the “Fluigent-MFCSTM-EZ” pressure control system. DNA amplification took place using the Eppendorf Mastercycler Gradient thermocycler. For optical readout, droplets were loaded into a disposable 10 µl cell counting chamber called “Countess™” without a grid, manufactured by EVETM NanoEnTek.
Image acquisition involved a standard fluorescence microscope (Axiovert-MAT-M, Carl Zeiss AG, Germany) equipped with a Zeiss Fluar 10x magnification NA 0.5 objective, HBO 100 light source, FITC-filter set, and an Andor-Neo sCMOS camera (Oxford Instruments, Abingdon, UK) with a 5-second exposure time for fluorescence images.
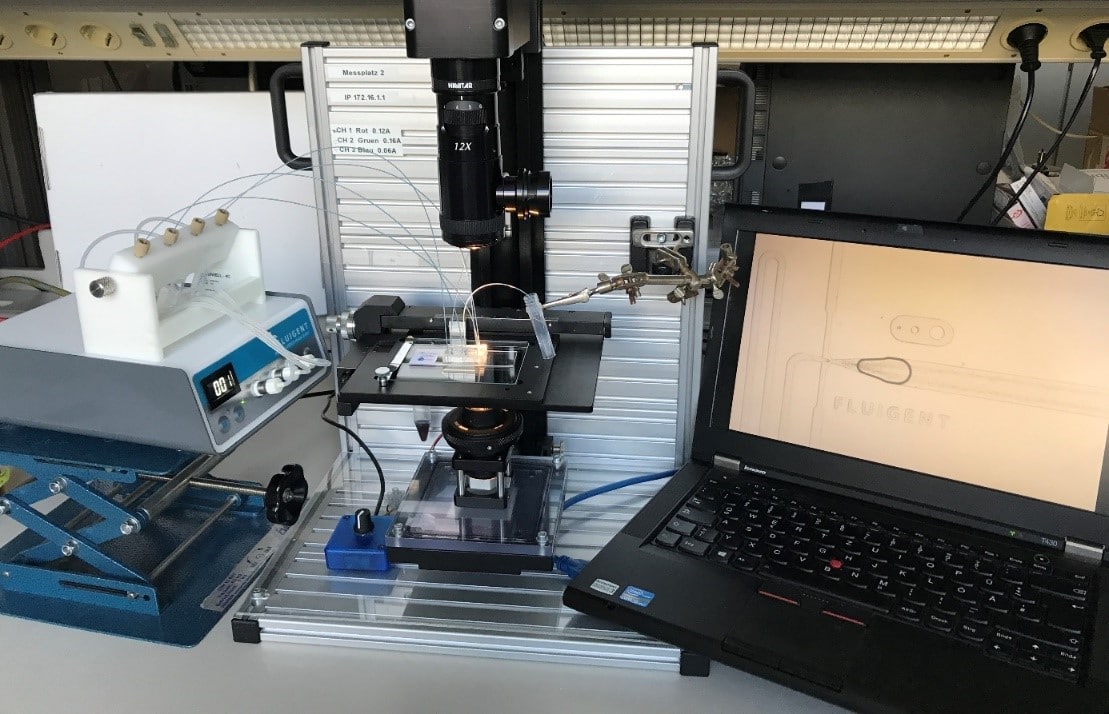
What is the method of dpcr?
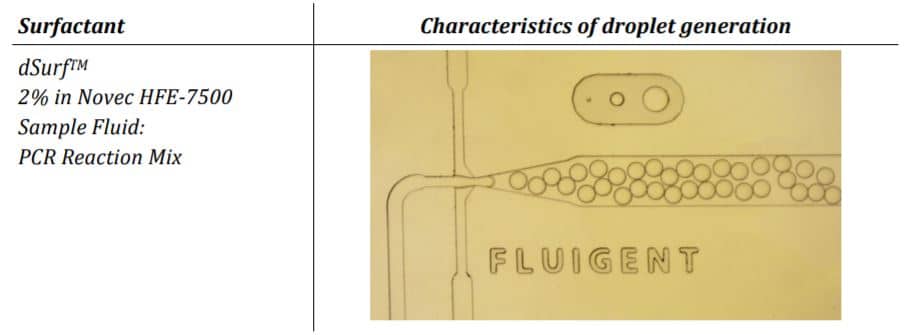
Droplets were generated at a working pressure of 240 mbar for the dSurf and 140 mbar for the PCR-Mix. The chip was connected with PEEK 1/32” tubing OD x .010” and 2x sleeves 1/16” OD x.033” ID x 1.6”, tubing length: 200 mm. Generated droplets were collected into a 0.2 ml PCR vial. Amplification was performed in a conventional thermocycler with the following settings:

To acquire images, the amplified droplets were transferred to a cell counting chamber for brightfield and fluorescence imaging. The droplets needed to be arranged as a monolayer within the chamber for effective readout. This was achieved by loading 10 µl of the droplet suspension into a pipette tip and allowing the droplets to rise. The entire volume was then loaded into the chamber, starting with the pure fraction of the continuous phase to ensure the droplets were injected into the partially pre-filled chamber. After loading, the rear slit of the chamber was sealed with adhesive tape to minimize evaporation and prevent droplet motion or rearrangement during the readout process.
dPCR assay data analysis
The parameter settings of the Fluigent-MFCSTM-EZ pressure control system, as described in the Materials and Methods section, were used to generate droplets for dPCR samples. Figure 2 illustrates the observed characteristics of the generated surfactant droplets, including the droplet generation regime, size, and frequency. The average droplet size was measured to be 70 µm, with a volume of 180 pL.
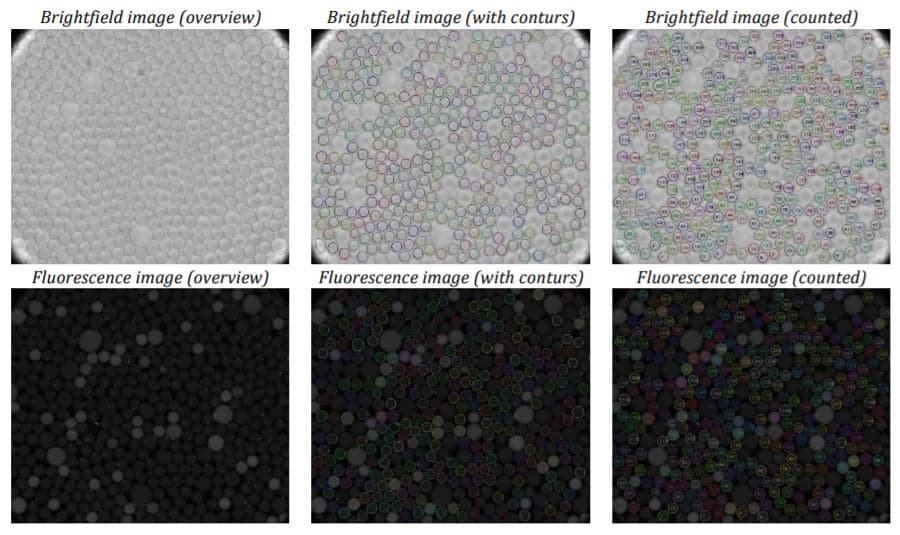
Conclusion
The experiments have shown that the dSurf surfactant is suitable for scientific as well as routine digital PCR applications. The generated droplets were homogeneous in shape and size. Superior droplet stability of the dSurf surfactant system was observed during the amplification process. A few droplets have dissipated during the experiments, but this can be neglected.
The reproducibility of the experiments was also confirmed. Droplet generation with identical parameters leads to identical droplet size and quality. Summarily, dSurf can be employed as a surfactant composition for digital droplet-based assays, and therefore, for digital PCR assay.
References
1. Pohl, G. and I.-M. Shih, Principle and applications of digital PCR. Expert review of molecular diagnostics, 2004. 4(1): p. 41-47.
2. Huggett, J.F., S. Cowen, and C.A. Foy, Considerations for digital PCR as an accurate molecular diagnostic tool. Clinical chemistry, 2015. 61(1): p. 79-88.
3. Quan, P.-L., M. Sauzade, and E. Brouzes, dPCR: a technology review. Sensors, 2018. 18(4): p. 1271.
Related Resources
- Microfluidics case studies
Using dSurf for High Throughput Laser-Induced Fluorescence Droplet Micro-Thermometry (LuMIn)
Discover - Interviews & Testimonials
Testimonials dSurf
Discover - Microfluidic Application Notes
E. Coli Culture in Droplets Using dSURF Fluorosurfactant
Discover - Microfluidic Application Notes
Microbiome culture in droplet using dsurf surfactant
Discover - Expertise videos
MICROFLUIDICS in DROPLET DIGITAL PCR
Discover - Microfluidic Application Notes
High-throughput cell DNA screening using digital PCR
Discover 
Droplet Digital PCR (ddPCR)
Discover


