Microfluidic chips: key applications
The principles of microfluidics have been applied in a wide variety of areas, including medical diagnostics, drug discovery, drug delivery, microscale chemical production, combinatorial synthesis and assays, artificial organs, microscale energy systems, and environmental sensing. Here we discuss some applications where microfluidics achieves results that would be very challenging to obtain when using conventional methods.
I. Droplet-based microfluidics
Microfluidic generation of droplets has attracted a lot of interest, enabling high throughput experiments by generating millions of micro-reactors and chambers in a few seconds. This method produces highly monodispersed droplets of very small volumes (μL to fL) of fluids with high frequency (up to hundreds of kHz), providing better control of processes like mixing, encapsulation, sorting, and sensing. Microfluidic-based droplets have many diverse and varied applications, such as particle synthesis3 and chemical analysis4. Highly controlled droplet production also enables single-cell analysis or drug testing 5,6.
Microfluidic-based droplet generation and control allow for:
- Highly monodispersed (<2% size variation) droplet production, with potentially high generation rate (up to hundreds of kHz)
- Highly reproducible complex structures (water-in-oil-in-water emulsions, multiple encapsulations…)
- Single droplet manipulation as an individual pL scale biochemical reactor
- Miniaturization of production and bioanalytical devices
With these characteristics, droplet microfluidics has a large value, including bio(chemical) analysis, and nano and microscale generation of materials7. Several chip designs exist to generate droplets. A common design is the T-junction, where the dispersed phase is injected from a channel that is perpendicular to the channel carrying the continuous phase (figure 1).
Droplet microfluidic concepts, components, and processes are now being adopted and leveraged by end-users to enable new science and innovation. Real-world success is now evidenced through a range of mainstream commercial products that are applied to key biological and healthcare-related problems (e.g., 10X Genomics, Drop-seq, and nucleic acid quantification via Droplet DigitalTM PCR systems)8.
Today, it is possible to produce multiple emulsions with complex droplet morphologies. Production of multi-cored droplets (droplets that contain a controlled number of inner droplets at one or more hierarchical levels), Janus droplets (i.e. biphasic or triphasic droplets with two or three physically and chemically distinct surface domains) is now possible using droplet microfluidics9 (figure 2).
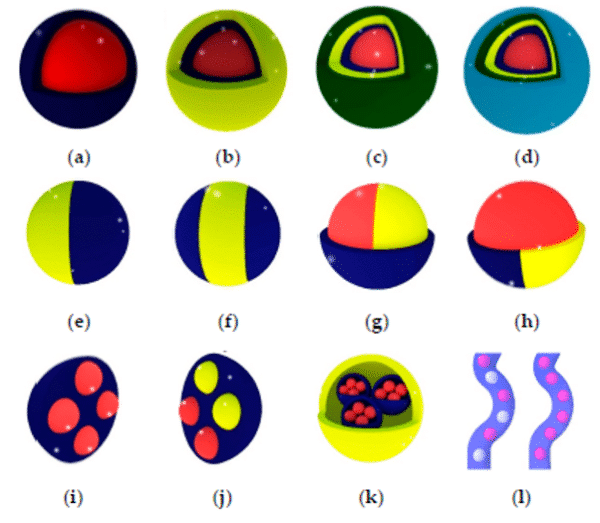
Figure 2: Types of multiple emulsions from the simple encapsulation (a), double emulsion (b), and so on (c,d) to bi- and tri-phasic structures (e to h), multiple encapsulations (I to k), and hybrid microjet achievable using droplet-based microfluidics9
A microfluidic chip for the production of highly reproducible PLGA microparticles
In recent years, biodegradable microspheres/microparticles have gained widespread importance in the delivery of bioactive agents10. The copolymers of Poly (D, L-lactic-co-glycolic acid) (called PLGA)/poly (lactic acid) (called PLA) microparticles are one of the most successful new drug delivery systems (DDS) in labs and clinics. Because of their biocompatibility and biodegradability, they can be used in various areas, such as long-term release systems, vaccine adjuvant, and tissue engineering11.
In PLGA microparticle production for drug release and delivery, microparticle size is a key parameter as it is directly related to the microparticles degradation rate and the drug release rate12. Although PLGA microparticle synthesis appears to be a successful drug delivery system, the current processes and tools to produce PLGA microparticles have many limitations, such as wide microparticle size distribution, poor repeatability, and aggressive chemical preparation conditions11. To solve these problems, droplet-based microfluidics offers an efficient method for improvement.
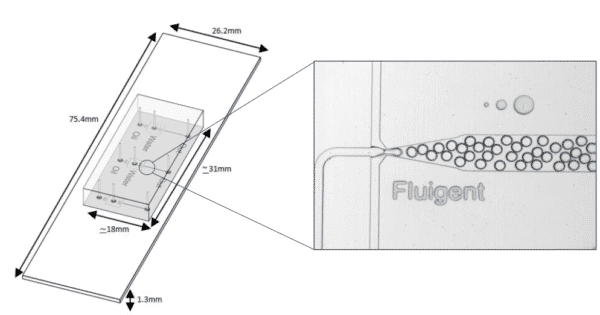
Fluigent provides the Raydrop: a new breakthrough technology leading to outstanding particle size monodispersity and production flexibility. The Raydrop works as a co-flow focusing principle (figure 3). The nozzle and outlet capillary are aligned in a continuous phase chamber, the dispersed phase comes through the nozzle to create the microparticles into the continuous phase and exits by the outlet insert. Using this system, PLGA microparticles ranging from 15 to 50 µm diameters have been successfully generated.
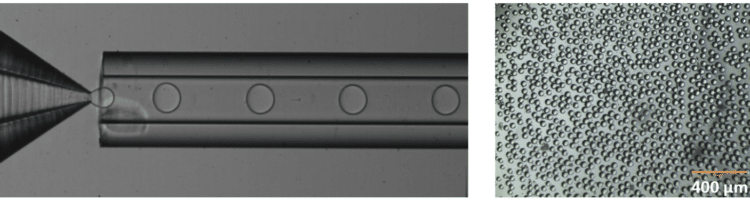
The PLGA microparticle production station allows excellent reproducibility and significantly improved monodispersity (CV < 2%) as compared to other methods on the market. It allows one to continuously produce PLGA microparticles (up to 10 000 droplets/s) without unwanted interruption for long-term experiments.
Single-cell mRNA-seq sequencing using droplets: Drop-seq technology
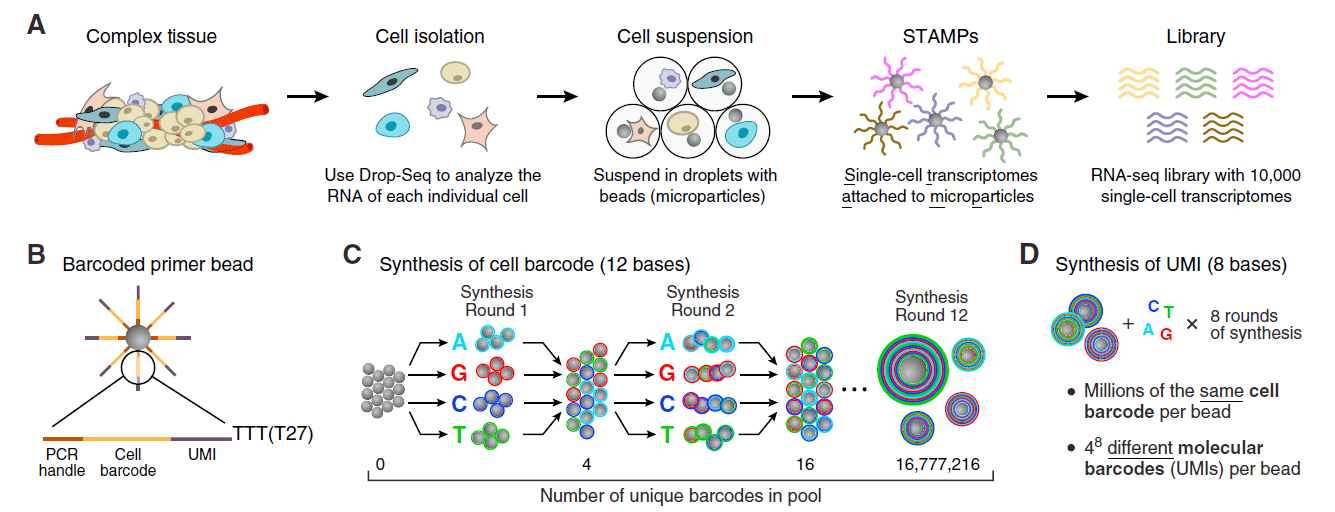
The production of highly monodispersed emulsions or more complex structures (water-in-oil-in-water emulsions, multiple encapsulations…) makes microfluidics an excellent tool for single-cell analysis or single-cell culture. The technique allows for droplet-based single-cell RNA-sequencing, such that one can characterize complex tissues with many cell types and states under diverse conditions. One of the pioneering methods is Drop-Seq technology, which entraps a single cell and a single primer-barcoded bead in each droplet (figure 4).
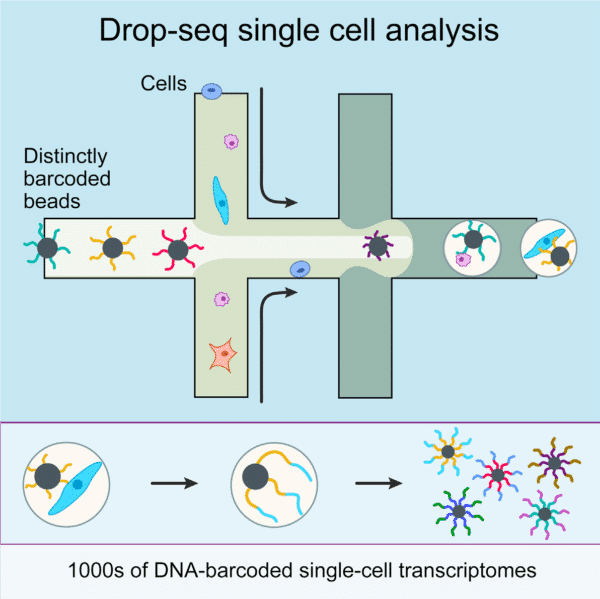
Cells are thus separated into nanoliter-sized aqueous droplets, with a different barcode associated with each cell’s mRNAs. The primers on beads contain a barcode consisting of three sequences. One sequence is for PCR amplification and is common to all the beads. The second sequence consists of hundreds of individual primers that also share the same ‘‘cell barcode’’. Finally, the third part has different unique molecular identifiers (UMI), enabling mRNA transcripts to be digitally counted13 (figure 5). The droplets are sequenced altogether, allowing quick profiling of thousands of individual cells from a heterogeneous population. The power of this technology resides in the fact that during sequencing, one can distinguish where the original information came on a cell to cell basis. This allows one to make a gene expression map of the cell, or even to distinguish cell populations within a tissue.
II. Microfluidic cell culture
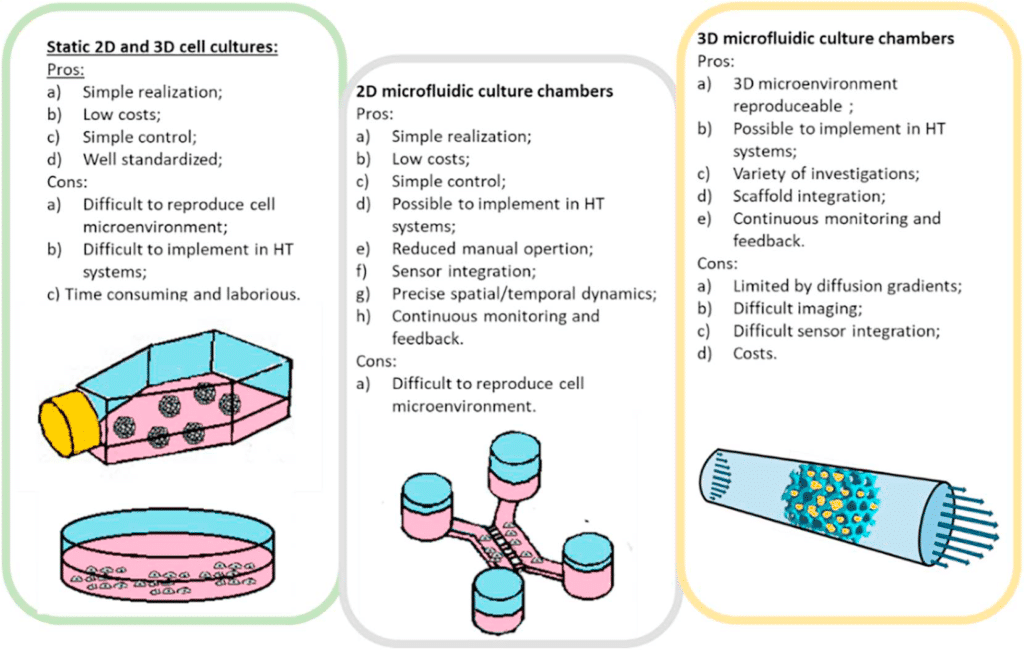
Microfluidic cell culture has significant advantages over macroscopic culture in flasks, dishes, and well-plates14 (figure 6). The microfluidic chip fabrication process allows great flexibility in the design of microfluidic devices, permitting one to understand and control interactions between cells, substrates, and the surrounding medium, physically as well as biochemically15.
This technology offers new possibilities to accurately reproduce the cellular environment and enables the analysis of biological processes that were not accessible before. Morphology-wise, chips can be structured at the cell scale to reproduce the mechanical constraints experienced by cells. Biochemically, stable gradients can be implemented with a high spatial resolution (typically, micrometer resolution).
Finally, constant perfusion enables the continuous renewal of nutrients and oxygen to promote cell growth and maintain optimal activity during long-term cell culture. Cost reduction due to volume reduction is also a major benefit15.
Development and characterization of a microfluidic model of the tumor microenvironment
The physical microenvironment of tumors is characterized by heterotypic cell interactions and physiological gradients of nutrients, waste products, and oxygen. This tumor microenvironment has a major impact on the biology of cancer cells and their response to chemotherapeutic agents. Despite this, most in vitro cancer research still relies primarily on cells grown in 2D and in isolation in nutrient and oxygen-rich conditions. Ayuso et al. presented an easy to use microfluidic device that can mimic the three-dimensional architecture of multicellular spheroids, while at the same time generating a visible, live “tumor slice” that allows easy monitoring of cells in different regions of the microenvironment in real-time as well as their response to different drugs17 (figure 7).
In this study, tumor cell behavior in different regions of the microdevice was studied and analyzed in conjugation with measurements of hypoxia and glucose concentrations across the device. The differential cellular response to several well-known drugs in different parts of the microdevice emphasizes the potential of this technology for analyzing the impact of microenvironmental parameters on drug response.
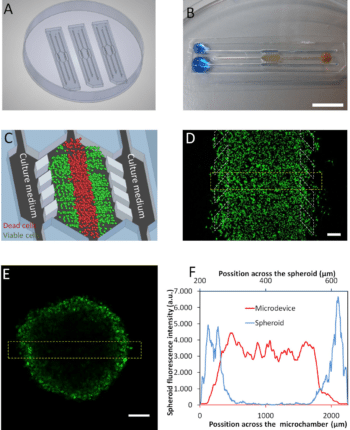
Figure 7: Microdevice operation of the tumor microenvironment. (A) 3 microdevices in a Petri dish containing a central culture chamber (detailed in C) and 6 channels. (B) One microdevice is filled with (yellowish) collagen hydrogel flowing to the microchamber from the right middle channel and blue-colored water perfused through the two lateral microchannels. (C) Culture medium perfused through the lateral microchannels provides nutrients and oxygen creating physiological gradients across the device. Cells near the ‘surrogate’ blood vessels are viable, whereas oxygen-poor cells in the center of device start to die creating a ‘necrotic core’ similar to the necrotic regions of tumors. (D) Cellular monitoring with fluorescent dye in the microdevice17.
III. Organ on a chip
Many efforts are devoted to the development of cancer metastasis models that can help in understanding the disease and the development of innovative therapeutic strategies. Current in vitro and in vivo cancer models are incapable of satisfactorily predicting the outcome of various clinical treatments on patients18. Therefore, new methods and approaches for drug discovery and health research are being developed. The concept of mimicking the organ-level function of human physiology or disease using cells inside a microfluidic chip was first published in 2004. In 2010 that the term organ-on-a-chip (OOAC) was invented by Ingber, et. al., who developed a microfluidic chip to capture organ-level functions of the human lung19. Microfluidics enables one the unique ability to control the cellular microenvironment with high spatiotemporal precision and to present cells with mechanical and biochemical signals in a more physiologically relevant context19. The manipulation of the micro-liter volumes of liquids has made these models a platform where scaling, and dynamic crosstalk between cells can be achieved. Microfluidic chips can now use geometries and structures to permit the use of physiological length scales, concentration gradients, and the mechanical forces generated by fluid flow to mimic the in vivo microenvironment experienced by cells20. These biomimetic platforms overcome many drawbacks encountered with conventional tissue culture models. OOAC engineering has attracted enormous interest and attention from the pharmaceutical industry, regulatory agencies, and even national defense agencies. This is demonstrated by the increase of OOAC research papers and by the emergence of at least 28 organ-on-a-chip companies in less than seven years21.
A microfluidic chip to reconstitue organ-level lung functions
To demonstrate that it is possible to engineer a microsystem that replicates the complex physiological functionality of living organs, Huh et al. developed a multifunctional microdevice that reproduces key structural, functional, and mechanical properties of the human alveolar-capillary interface, which is the fundamental functional unit of the living lung19. The device consisted of compartmentalized PDMS microchannels to form an alveolar-capillary barrier on a thin, porous, flexible PDMS membrane coated with the extracellular matrix and human alveolar epithelial cells and human pulmonary microvascular endothelial cells is cultured on opposite sides of the membrane (figure 8). Air is subsequently introduced into the compartment to create an air-liquid interface to mimic the lining of the alveolar air space19
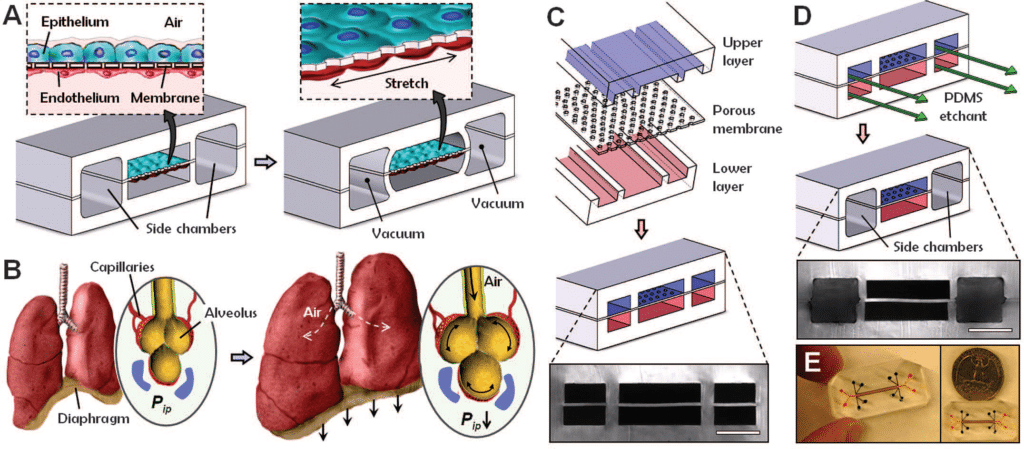
Using this organ on a chip platform, the authors demonstrated that breathing motions, simulated by the platform, might greatly accentuate the proinflammatory activities of silica nanoparticles and contribute substantially to the development of acute lung inflammation19. This behavior could not be determined using existing in vitro models.
IV. Particle/cell sorting
Isolating and sorting cells from complex, heterogeneous mixtures represents a critical task in many areas of biology, biotechnology, and medicine. Cell sorting is often used to enrich or purify cell samples into well-defined populations to enhance efficiency in research and development applications22. Cell detection is generally performed using optical methods such as FACS (Fluorescent Activated Cell Sorting), where cells pass through a laser beam and the light scattered is characteristic to the cells and their components. Cells are subsequently sorted according to the scattered emission. It is an automated and robust solution for cell sorting and was considered a gold standard for cell sorting. The immense contributions of current commercial cell sorting platforms using FACS are tempered by several significant and persistent limitations, including limited sample throughput and processing speeds that make the generation of clinical-scale samples very difficult.
As opposed to conventional instrumentation, microfluidic chips are affordable, easy to use, and smaller. These chips make use of a wide range of techniques to sort cells, with specific speeds and efficiencies. The dimensions of the chip also allow a wide range of cell sorters to exist, from large volume cell sorters to precise single-cell sorters. Moreover, microfluidic cell sorting can be combined with additional fluidic operations within a single chip for complete lab-on-a-chip applications, diagnostic and therapeutic purposes. Microfluidic cell sorting is a promising application both in academic and industrial labs. Cell sorting is based on determining a specific cell parameter that can differ from one cell type to another, such as cell size, shape, density, surface markers. In general, a heterogeneous cell solution is injected through a microfluidic chip.
The chip is designed such that cells with different properties experience a different amount of force (that could originate from inertia, channel geometry, external sources …) while similar cells undergo equal forces. The differentiated cells are subsequently pushed into different streamlines and exit the chip from different outlets. Microfluidic cell sorting can be categorized into three distinct groups: fluorescent label-based, bead-based, and label-free cell sorting22. Label-free cell sorting in microfluidic devices is perhaps the most studied of the three approaches as it encompasses active systems (i.e., systems that rely on the use of external fields cells for sorting), and passive systems that do not rely on fluorescent labels or beads. Instead, these methods rely on the inherent differences in cellular morphology between cell groups22.
Inertial microfluidics for continuous particle separation in spiral microchannels
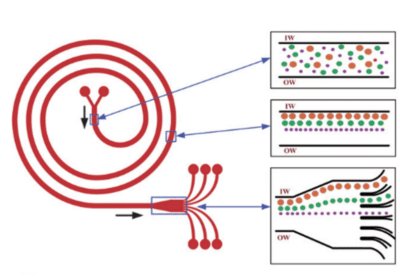
Kuntaegowdanahalli et al. developed a simple inertial microfluidic device that achieves continuous multi-particle separation using the principle of Dean-coupled inertial migration in spiral microchannels.
The dominant inertial forces coupled with the Dean rotational force due to the curvilinear microchannel geometry, cause particles to occupy a single equilibrium position near the inner microchannel wall. The position at which particles equilibrate is dependent on the ratio of the inertial lift to Dean drag forces.
Using this concept, they demonstrated a spiral lab-on-a-chip for size-dependent focusing of particles at distinct equilibrium positions across the microchannel cross-section from a multi-particle mixture23.
A major benefit of this system is its high throughput (e.g., 1.5 mL/min) without sheath flow or sequential cell manipulation, which is useful for processing native biological fluids and flow cytometry.
V. Micromixers
Micromixers are important components of lab-on-chip devices for applications in drug delivery, sequencing, amplification, and biochemical reactions. Based on the actuation method, micromixers can be broadly classified as passive and active. In passive mixing, no external sources are used. Thus mixing typically relies on the microfluidic chip geometry and on fluid properties. Under laminar flow, which is the typical fluid regime in microfluidics, mixing mostly happens through diffusion. This property allows performing mixing using lamination: two or more liquids are flowing in parallel, allowing for diffusion to happen and thus permitting to precisely tune mixing between liquids. For improved and faster mixing, chaotic advection can be generated by modifying the microfluidic chip geometry, such as by altering the shape of the channel for splitting, folding, stretching, or breaking the flow of fluid.
Active mixing occurs by using an external perturbation. Many active mixing methods exist. Dielectrophoresis mixing makes use of an electric field to move particles toward or away from an electrode, creating a chaotic advection. Acoustic wave energy can also be used to mix fluids: strong acoustic waves can be produced, and interfere with each other as they propagate in the fluid, generating advection24. As the diffusion coefficient of a liquid depends on the temperature, mixing can also be enhanced by increasing the microfluidic chamber temperature25.
Submillisecond organic synthesis using a serpentine-shaped microfluidic chip
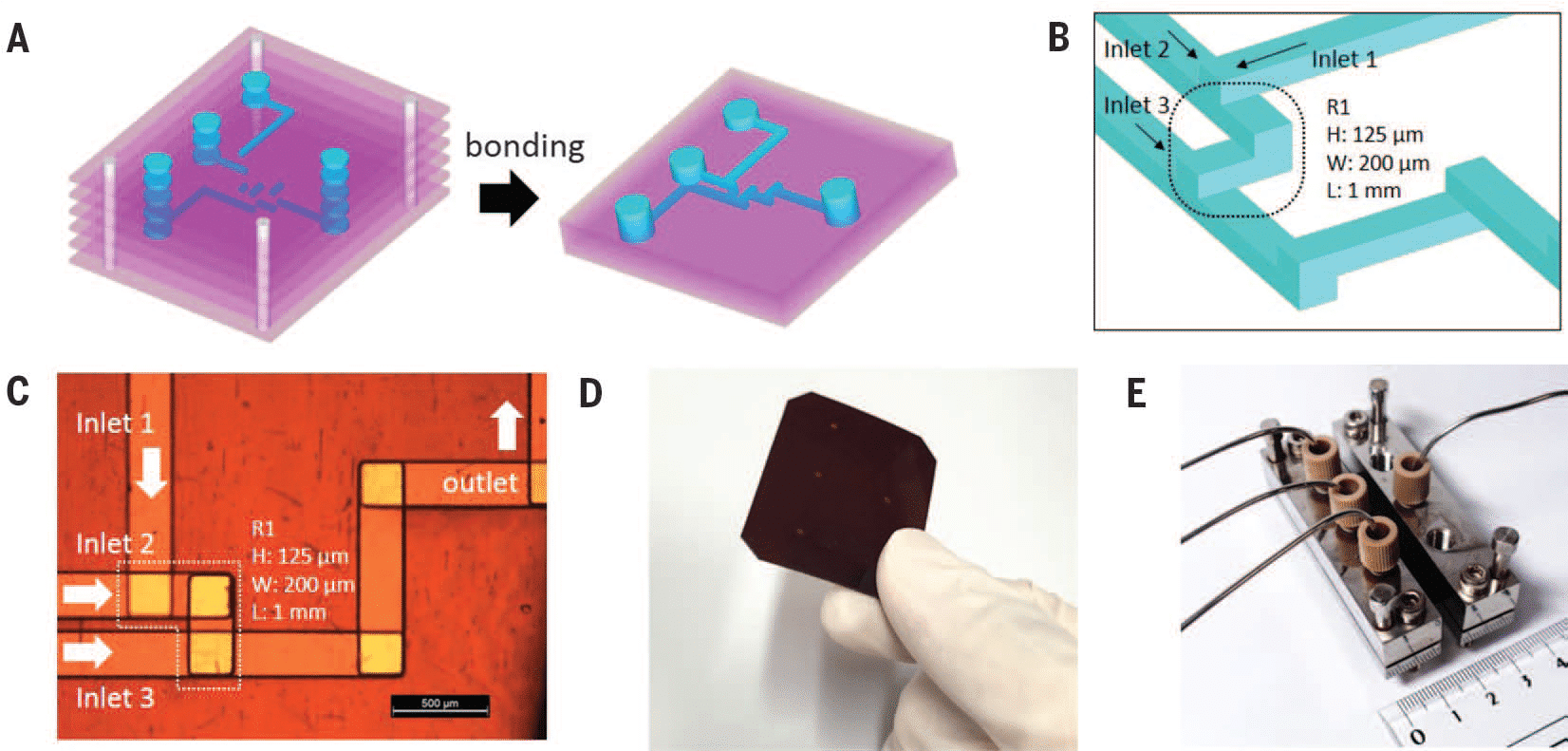
In chemical synthesis, it is important to explore the synthetic pathways of an intermediate. To fully observe these pathways, control over its lifetime and mixing time is required.
The reaction mixture had to be well-mixed within the lifetime of the reactive intermediate. An efficient means of prolonging the lifetime is to lower the reaction temperature (-78°C to -100 °C), above the melting point of many organic solvents. Using microfluidic devices, mixing can be extremely fast, with mixing times unattainable by batches26. However, mixing time is increased at low temperatures, as the solvent viscosity exponentially increases.
To circumvent this issue, the authors used a three-dimensional serpentine-shaped microfluidic chip, allowing improved mixing by chaotic advection. Using this microfluidic chip, submillisecond mixing was performed.
VI. Microvalves
In the past 10 years, efforts have been devoted to the development of microfluidic platforms capable of performing several assays using programmable fluidic operations within an array of microvalves. Similar to programmable logic circuits where multiple electronic computing routines are executed on a single microdevice, programmable microfluidic platforms have been implemented27, allowing one to perform fluidic operations such as mixing, sampling, washing, and reacting automatically within a single chip by modifying the sequence/order of fluidic operations using the software. The primary advantage of microvalves over their macroscale counterparts is the significantly reduced dead volume, which is important in many applications that require precise flow control at small flow rates28. They are useful in biological and chemical applications, such as quantitative metabolic biomarker and genetic analysis29,30, protein-based biomarker detection31, or small molecule chemical and environmental analysis32. These platforms usually consist of a 2D array of microvalves that permit flow regulation, on/off switching and sealing of liquids, gases, or vacuums33. Several microvalves have been developed using pneumatic, electrokinetic and electrochemical actuators. Among these mechanisms, pneumatic actuation is often recognized as the most reliable method due to the simplicity of fabrication, ease of use, scalability, reliability, and a high degree of accuracy. Pneumatically actuated microvalves utilize the deflection of an elastomer (typically PDMS) membrane to control fluid flow34.
A fully integrated multilayer microfluidic chemical analyze for automated sample processing, labelling, and analysis
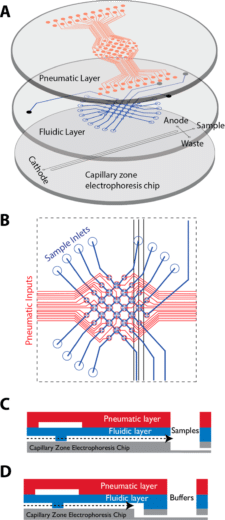
Capillary zone electrophoresis (CZE), is a powerful tool for chemical analysis and is widely used for environmental monitoring, astrobiology, and biosensing32. CZE assays usually require complex and manual sample processing.
Commercial platforms for automated CZE have been implemented to address this concern, but are expensive, and large, thus hardly field deployable in challenging environments. Microfluidic systems allow miniaturization, automation, and reduction in sample volume requirements for chemical and biochemical sensing.
Using membrane microvalve technology, it is possible to automate metering, transporting, routing, and mixing operations. Kim et al. introduced a microfluidic platform consisting of pneumatically actuated “lifting gate” microvalves integrated with a glass CZE microchip, providing extremely low dead volumes between components.
All the procedures, including buffer filling, labeling, and dilution, can be automated. The microchip was used to analyze diverse compound classes, such as amino acids, and oxidized biomarker compounds, like aldehydes/ketones and carboxylic acids in less than 30 min.
VII. Wearable microfluidics: an emerging application
An emerging field is the use of microfluidic concepts for wearable device applications36. Here, microfluidics presents several key value propositions. The microstructures store or handle fluids and are the core of the sensing device. Using microchannels, precise liquid amounts can be manipulated, allowing for highly accurate and reliable devices and being useful for bodily fluids that are often secreted or extracted in limited quantities. Also, wearable microfluidic devices could also stock a specific drug for precise dispensing at controlled intervals. Innovations in flexible microfluidics and electronics have led to numerous applications. Typically, a wearable microfluidic device will collect a fluid, transfer it to a localized site where detection or measurement is performed. Blood and sweat are common analytes as they provide insights into physiological states such as temperature, pH, and hydration36. Wearable microfluidics finds applications in the pharmaceutical, food, sportswear, and cosmetic fields.
A wearable microfluidic device for the capture, storage and colorimetric sensing of sweat

As mentioned previously, sweat is an analyte of interest because of its rich content of important biomarkers. It is easy to collect compared to blood. In situ quantitative analysis of sweat is of great interest for monitoring physiologic health status (for example, hydration state) and for the diagnosis of disease37.
Existing systems for sweat collection and analysis are confined to laboratories, where standard analytical technologies can be performed. Though highly precise, the analysis is time-consuming and costly.
To address this issue, Kho et al. developed a soft wearable microfluidic system than can directly harvest sweat from pores on the surface of the skin37.
The device routes the sample to different channels and reservoirs for multiparametric sensing of markers of interest, with options for wireless interfaces to external devices for image capture and analysis.
The device can measure total sweat loss, pH, lactate, chloride, and glucose concentrations by colorimetric detection using wireless data transmission. As it is a simple, low-cost, and fast analysis point of care device, it could be used to accumulate data from individual users over time, and this could serve as an analytical approach for interpreting trends in marker concentrations, potentially providing warning signs when performing physical activity.
Conclusion
Since the introduction of microfluidics, the scope of applications has kept extending over the years. The first applications were focused on analytical chemistry, but today the field of life science and specifically point of care is in the core of microfluidics. We have presented applications where microfluidics show great advantages compared to conventional systems. The importance of these applications was illustrated by showing research papers related to these applications. Some important applications were introduced here. Microfluidics covers a wide range of applications such as microreactors, bioprinting, or fuel cells, and many more.
References
- Seemann, R., Brinkmann, M., Pfohl, T. & Herminghaus, S. Droplet based microfluidics. Reports Prog. Phys. 75, (2012).
- Paquin, F., Rivnay, J., Salleo, A., Stingelin, N. & Silva, C. Droplet Control Technologies for Microfluidic High Throughput Screening (µHTS). Muhsincan Sesen,a Tuncay Alan,a and Adrian Neild∗a 10715–10722 (2017) doi:10.1039/b000000x.
- Galas, J. C., Bartolo, D. & Studer, V. Active connectors for microfluidic drops on demand. New J. Phys. 11, (2009).
- Jullien, M.-C., Tsang Mui Ching, M.-J., Cohen, C., Menetrier, L. & Tabeling, P. Droplet break in a low capillary T-junction. in 19th Mechanical French Congress (AFM, Maison de la Mécanique, 39/41 rue Louis Blanc-92400 Courbevoie, 2009).
- Yu, L., Chen, M. C. W. & Cheung, K. C. 2010 Droplet-based microfluidic system for multicellular tumor spheroid formation and anticancer drug testing. Lab Chip 10, 2424–2432 (2010).
- N.Shembekar, C.Chaipan, R. U. & C. A. M. 2016 Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics. Lab Chip (2016) doi:10.1039/C6LC00249H.
- Shang, L., Cheng, Y. & Zhao, Y. Emerging Droplet Microfluidics. Chem. Rev. 117, 7964–8040 (2017).
- Suea-Ngam, A., Howes, P. D., Srisa-Art, M. & Demello, A. J. Droplet microfluidics: From proof-of-concept to real-world utility? Chem. Commun. 55, 9895–9903 (2019).
- Vladisavljević, G. T., Al Nuumani, R. & Nabavi, S. A. Microfluidic production of multiple emulsions. Micromachines 8, (2017).
- Soppimath, K. S. & Aminabhavi, T. M. Ethyl acetate as a dispersing solvent in the production of poly(DL-lactide-co-glycolide) microspheres: Effect of process parameters and polymer type. J. Microencapsul. 19, 281–292 (2002).
- Qi, F., Wu, J., Li, H. & Ma, G. Recent research and development of PLGA / PLA microspheres / nanoparticles : A review in scienti fi c and industrial aspects. (2018).
- Anderson, J. M. & Shive, M. S. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv. Drug Deliv. Rev. 64, 72–82 (2012).
- Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
- Halldorsson, S., Lucumi, E., Gómez-Sjöberg, R. & Fleming, R. M. T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 63, 218–231 (2015).
- Yeo, L. Y., Chang, H. C., Chan, P. P. Y. & Friend, J. R. Microfluidic devices for bioapplications. Small 7, 12–48 (2011).
- Coluccio, M. L. et al. Microfluidic platforms for cell cultures and investigations. Microelectron. Eng. 208, 14–28 (2019).
- Ayuso, J. M. et al. Development and characterization of a microfluidic model of the tumour microenvironment. Sci. Rep. 6, 1–16 (2016).
- Caballero, D. et al. Organ-on-chip models of cancer metastasis for future personalized medicine: From chip to the patient. Biomaterials 149, 98–115 (2017).
- Huh, D. et al. Reconstituting organ-level lung functions on a chip. Science (80-. ). 328, 1662–1668 (2010).
- Bhise, N. S. et al. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Release 190, 82–93 (2014).
- Zhang, B., Korolj, A., Lai, B. F. L. & Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 3, 257–278 (2018).
- C. Wyatt Shields IV, Dr. Catherine D. Reyes, and P. G. P. L. Microfluidic Cell Sorting: A Review of the Advances in the Separation of Cells from Debulking to Rare Cell Isolation. Lab Chip (2015) doi:10.1039/c4lc01246a.
- Kuntaegowdanahalli, S. S., Bhagat, A. A. S., Kumar, G. & Papautsky, I. Inertial microfluidics for continuous particle separation in spiral microchannels. Lab Chip 9, 2973–2980 (2009).
- Vivek, V., Zeng, Y. & Kim, E. S. NOVEL ACOUSTIC-WAVE MICROMIXER.
- Ward, K. & Fan, Z. H. Mixing in microfluidic devices and enhancement methods. J. Micromechanics Microengineering 25, (2015).
- Kim, H. et al. Submillisecond organic synthesis: Outpacing Fries rearrangement through microfluidic rapid mixing. Science (80-. ). 352, 691–694 (2016).
- Thorsen, T., Maerkl, S. J. & Quake, S. R. Microfluidic large-scale integration. Science (80-. ). 298, 580–584 (2002).
- Aditya Aryasomayajula, Pouriya Bayat, Pouya Rezai, P. R. S. Microfluidic Devices and Their Applications. Springer vol. 50 (2017).
- Jensen, E. C., Bhat, B. P. & Mathies, R. A. A digital microfluidic platform for the automation of quantitative biomolecular assays. Lab Chip 10, 685–691 (2010).
- Vincent, M., Xu, Y. & Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 5, 795–800 (2004).
- Erik C. Jensen, Yong Zeng, Jungkyu Kim, and R. A. M. Microvalve Enabled Digital Microfluidic Systems for High Performance Biochemical and Genetic Analysis. 15, 455–463 (2011).
- Kim, J., Jensen, E. C., Stockton, A. M. & Mathies, R. A. Universal microfluidic automaton for autonomous sample processing: Application to the mars organic analyzer. Anal. Chem. 85, 7682–7688 (2013).
- Oh, K. W. & Ahn, C. H. A review of microvalves. J. Micromechanics Microengineering 16, (2006).
- Kim, J., Stockton, A. M., Jensen, E. C. & Mathies, R. A. Pneumatically actuated microvalve circuits for programmable automation of chemical and biochemical analysis. Lab Chip 16, 812–819 (2016).
- Chin, V. I. et al. Microfabricated platform for studying stem cell fates. Biotechnol. Bioeng. 88, 399–415 (2004).
- Yeo, J. C., Kenry & Lim, C. T. Emergence of microfluidic wearable technologies. Lab Chip 16, 4082–4090 (2016).
- Ahyeon Koh, Daeshik Kang, Yeguang Xue, Seungmin Lee, Rafal M. Pielak, Jeonghyun Kim, Taehwan Hwang, Seunghwan Min,1 Anthony Banks, Philippe Bastien, Megan C. Manco, Liang Wang, Kaitlyn R. Ammann, Kyung-In Jang, Phillip Won Seungyong Han, Roozbeh Ghaffari, J. A. R. A soft, wearable microfluidic device for the capture, storage, and colorimetric sensing of sweat. Sci. Transl. Med. 26, 39–46 (2017).